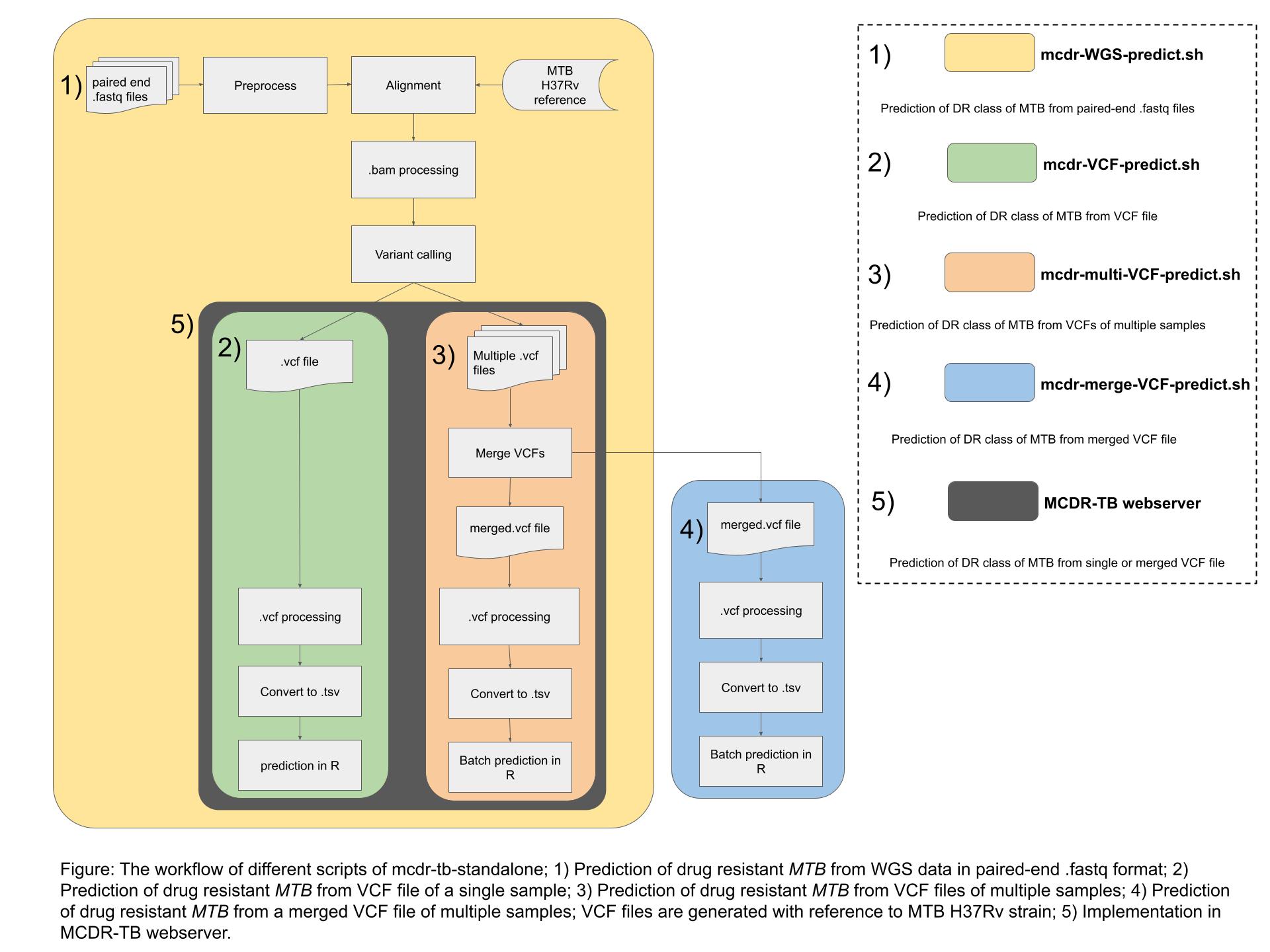
MCDR-MTB is a pipeline that uses whole genome sequencing data to categorize MTB isolates into three drug resistance classes.
Users can use MCDR-MTB webserver by uploading the Variant calling format (VCF) file of single MTB isolate or merged VCF file with more than one MTB isolates.
Users can generate VCF files using their own pipelines or they can use
mcdr-mtb-utilities scripts. Two scripts are provided - 1)
variant-call.sh - The script to perform variant calling of MTB paired-end WGS sequences with reference to H37Rv genome.
2)
merge-VCF.sh - The script to merge multiple VCF files into a single VCF file.
To predict drug resistance class of multiple MTB isolates, users can install
mcdr-mtb-standalone. The standalone version has scope to predict the class from raw sequences in FASTQ format as well as analysed VCF files. The
figure below shows the overall structure of MCDR-MTB webserver and standalone tool.