|
What is RHObase?
Several information regarding Rieske-type aromatic ring-hydroxylating oxygenases (RHOs) are available online in various public databases either in the form of sequence, structure, function or in literature. However, these information are scattered through different discrete information bases and it becomes difficult for researchers to navigate through all available information regarding this family of enzymes. ?RHObase?, an abbreviation for Ring-Hydroxylating Oxygenase database, is a web based, manually curated and searchable database which provides comprehensive information on Rieske-type RHOs on a single platform along with analytical tools to perform blast search against the database for prediction of putative substrate(s) for query oxygenase sequence(s) or to search for potential candidate RHOs capable of transforming a desired compound.
Go to top
What are RHOs?
RHOs are multi-component enzyme system involved in degradation of diverse aromatic compounds in the environment which include linked and fused aromatics, aliphatic olefins, and highly substituted aromatics as well as many toxic and/or carcinogenic compounds such as polychlorinated biphenyls (PCBs) and polycyclic aromatic hydrocarbons (PAHs). All members of the RHO family have one or two soluble electron transport (ET) proteins, viz. ferredoxins and reductases, having flavin and/or Fe-S centers, which transfer electrons from reduced nucleotides, NAD(P)H, to the terminal oxygenase component (Fig. 1).The terminal oxygenase mostly comprises of two separate subunits, a large catalytic subunit (α) with a Rieske-type Fe-S catalytic centre and a small structural subunit (β) in hetero-multimeric form, αβn. However, certain dioxygenases are devoid of β-subunit, and exist in homo-multimeric form, αn.
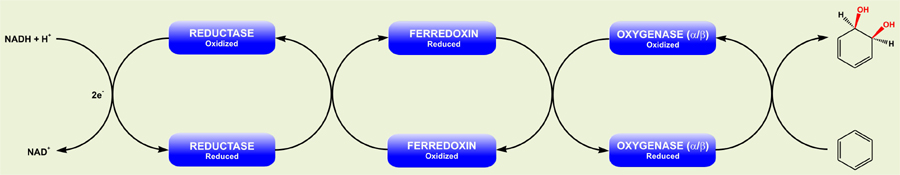
Figure 1
Go to top
Why focus on RHOs?
RHOs initiate the catabolism of several recalcitrant compounds by attacking the inert aromatic nucleus making them prone to further transformation and mineralization. These enzymes are thus crucial for studying degradation of different organo-pollutants. Moreover, the regio- and stereo-specific nature of catalysis by these enzymes make them ideal candidates for lead development and natural product modification from the standpoint of pharmaceutical and synthetic chemists.
Go to top
How users will be benefited from the RHObase?
RHObase will be helpful for a wide variety of researchers. This includes environmental biologists dealing with transformation of any candidate xenobiotic compound, synthetic chemists attempting to synthesize stereo-selective dihydrodiol drivatives as well as molecular biologists trying to predict the functional aspects of a de novo sequenced oxygenase (from whole genome or metagenome sequencing). Thus, RHObase provides all necessary information and prediction tools about RHOs which are required prior to designing an experiment.
Go to top
What information can I obtain from the database?
The current version of the database compiles ∼1000 entries including 185 oxygenase α-subunits, 142 oxygenases β-subunits, 90 ferredoxins and 108 reductases, distributed among 129 different bacterial strains. The proteins are linked to available PDB structures and corresponding conserved domains (and motifs). The database also includes information on more than hundred aromatic compounds and the oxygenation mechanisms followed by different RHOs. The database allows users to search a query based on organism name, oxygenase, substrate or protein structure. The data is organized in a hierarchical manner (Fig. 2) where you can retrieve information regarding its classification, primary substrate, reaction mechanism, structures, and constituent proteins of RHOs. In addition, you can get detail information about conserved domain(s) with motif signature. Furthermore, names of a wide range of aromatic compounds which can be degraded by RHOs are also available. The entries are also linked to several external databases, such as ExPASy, PDB, CDD, KEGG, PubChem, ChEBI and UMBBD for additional information, as well as to PubMed, for related reference.
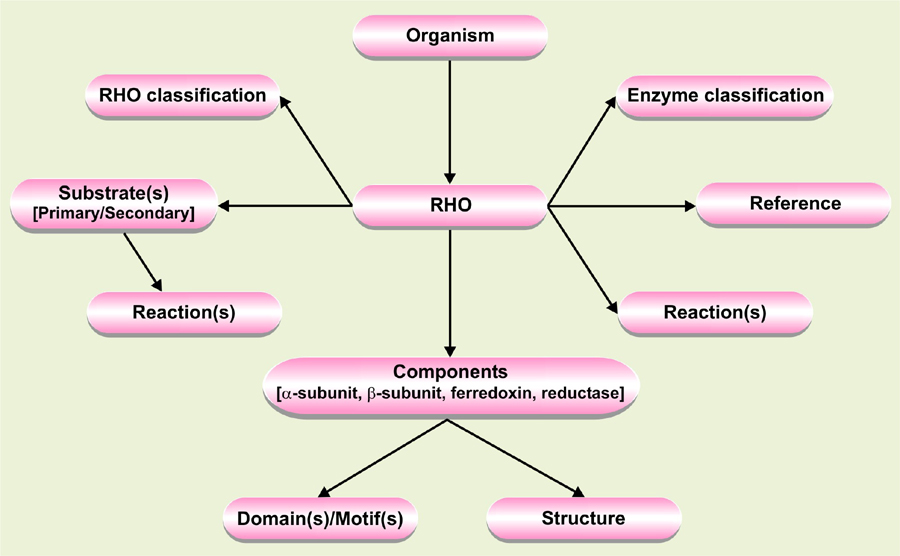
Figure 2
You can retrieve information in the form of five different pages relevant to your search query:
The Oxygenase page: When searched with a query string against a specific field (organism name, oxygenase or substrate), you will retrieve relevant information from the Oxygenase page. For a query genus, species or strain, the page displays information about all known RHO clusters present in the organism, along with links to RHObase ID of constituent components (α-subunit, β-subunit, ferredoxin and reductase), substrate(s) known to be transformed by the RHO(s) (with reaction mechanism) as well as corresponding PubMed link (Fig. 3). Similarly, for a query oxygenase (or substrate), you will get information regarding the organisms in which the RHO has been reported (or the organisms with their RHOs known to transform the substrate) and all other information as mentioned above.
[NB: The following teminologies may be encountered in certain result pages:
NA: data not available; Absent: the component is not required by the oxygenase; Unknown: no sequence/substrate information; Shared: the component (mostly electron transport) is shared from another RHO cluster within the same organism; None: does not exist.]
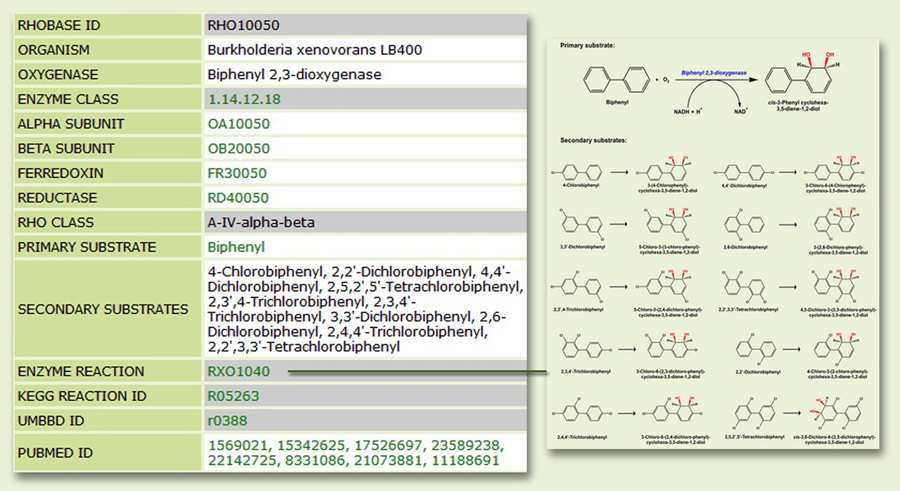
Figure 3
The Substrate page: This page bears information regarding the RHOs that are known to transform the substrate and their reaction mechanisms (Fig. 4). The page is also linked to compound databases such as KEGG, PubChem and ChEBI.
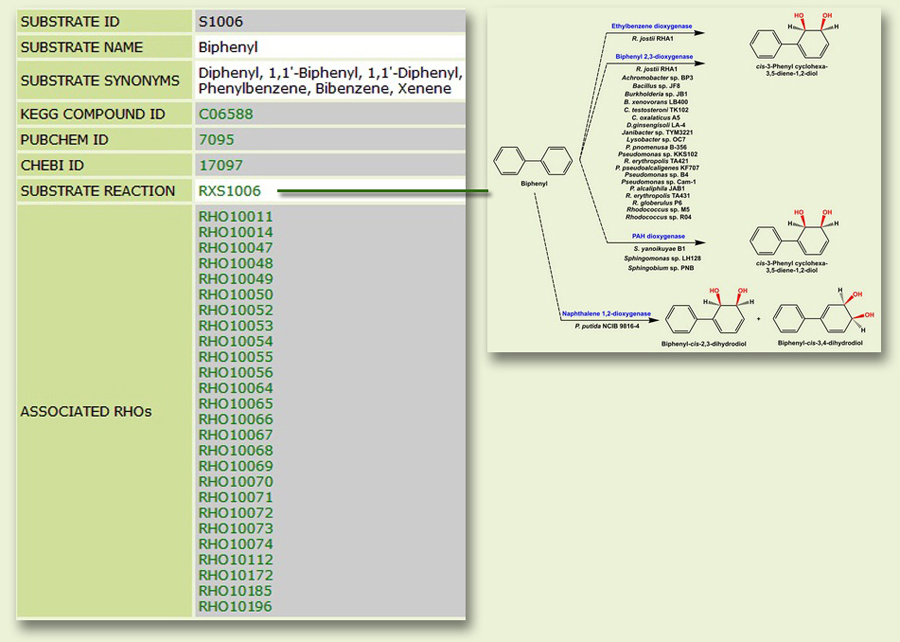
Figure 4
The Component page: This page displays information regarding the designated protein name of the selected component, NCBI accession number, PDB link to its 3D structure (if available) and conserved domain(s) (Fig. 5).
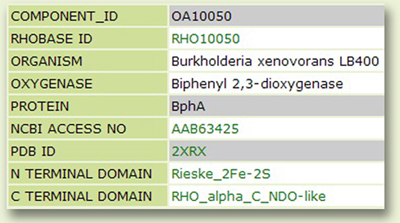
Figure 5
The Domain page: You will obtain detailed information about the selected domain along with its motif signature generated from sequence alignment of diverse members of the group (Fig. 6).
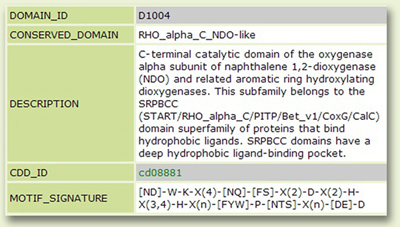
Figure 6
The Structure page: For a query structure, this page displays information about relevant PDB structures available for constituent components of the RHO(s) (Fig. 7).
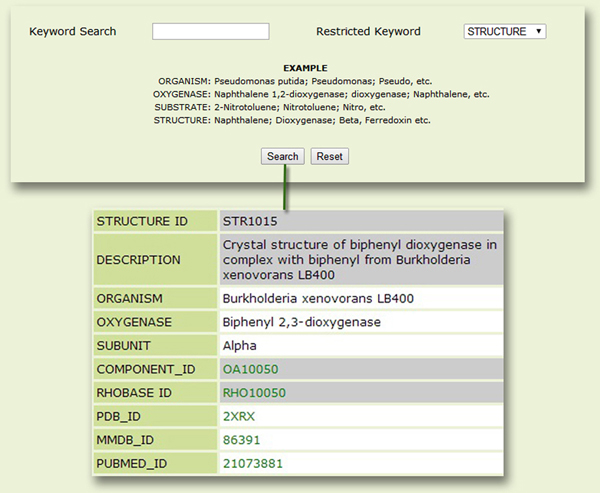
Figure 7
Go to top
How do I search for a query in the RHObase search page?
You can search the database with one of the five query types (Fig. 8):
ALL: Any string (such as organism name, oxygenase or substrate) may be submitted as a query under this field.
ORGANISM: Enter the genus name and/or the species name, or a strain name or any text string within the name of an organism.
OXYGENASE: Here you may search with the name of any oxygenase enzyme (e.g., "naphthalene 1,2-dioxygenase" or simply "naphthalene dioxygenase") or you may also search for different types of Rieske-type RHOs (like oxygenase, dioxygenase, monooxygenase, hydroxylase, demethylase, etc.)
SUBSTRATE: Enter the name of a desired substrate to retrieve the RHO(s) reported to transform that substrate.
STRUCTURE: Enter the name of a desired oxygenase or substrate to retrieve available PDB structures relevant to the query oxygenase/substrate.
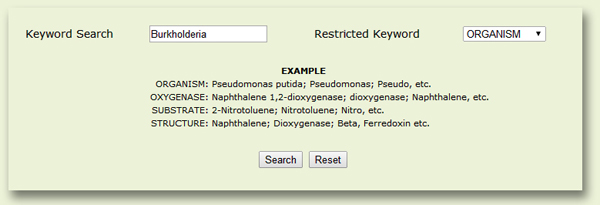
Figure 8
Go to top
I can't find my search item in the database, why?
If you did not find any result in the first hit, try searching with a sub-string of your query; e.g., if searching with the "2-nitrotoluene" does not give any result, try searching with "nitrotoluene" or "nitro" or "toluene". If none of these works, then the database possibly does not have the data you are searching for.
Go to top
What does “RHO_CLASSIFICATION” in the Oxygenase page mean?
RHOs are classified into 11 different types falling within 5 distinct classes (A, B, C, D and D*) taking into account both the functional evolution of catalytic Α-subunit as well as all possible combinations of electron-transport components (ferredoxin and reductase) associated with the terminal oxygenase. Such a classification reflects the evolutionary and functional aspects of RHOs. For further information, please refer to: Chakraborty et al. (2012). J. Biomol. Struct. Dyn. 30:419-36.
Go to top
How does the ‘Analysis‘ module work?
There are two analytical tools integrated with RHObase. The first one allows users to perform blast search (Fig. 9) against the database for prediction of putative substrate(s) for their query oxygenase sequence(s). Details of the blast analyses (best hit obtained for each component) and predicted putative substrate(s) for the query oxygenase are returned as results (Fig. 9). In addition, users can also view the sequence alignment (constructed using MultAlin) and phylogenetic tree (constructed using ClustalW and TreeExplorer)
of α-subunit protein sequences belonging to that group. One can also download the sequence file, append their own sequence and perform multiple sequence alignment (such as ClustalW).

Figure 9
The second prediction tool can be used to search for a query aromatic compound, either submitted directly as MOL file or drawn from the JME molecular editor (Fig. 10), for similar compounds in RHObase. The search result displays similar compounds (compound ID, linked to the database for further details) with corresponding Tanimoto similarity score.
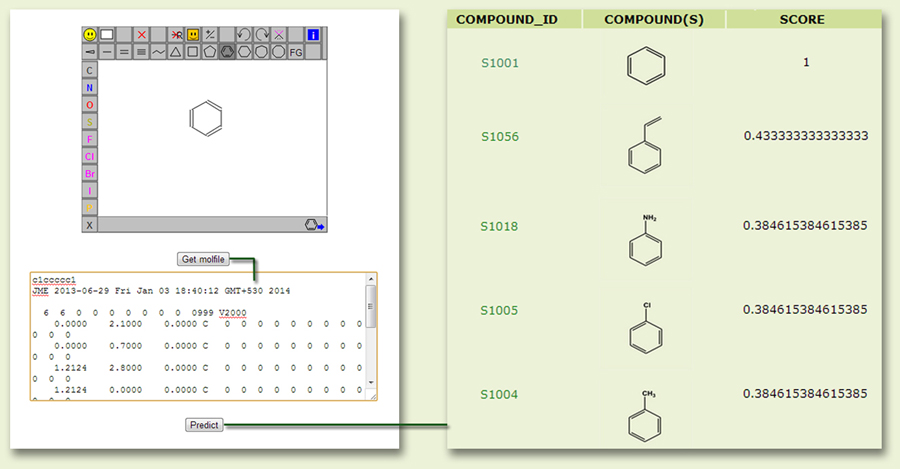
Figure 10
Go to top
How do I report a bug in RHObase?
Please email Dr. Sudipto Saha (ssaha4@jcbose.ac.in) with any bugs that you think need attention, with as much detail as possible, including the RHObase ID, nature of the bug and what needs to be done to fix it.
Go to top
My information in the database is not correct. How can I update my object(s)?
Please email Dr. Tapan K. Dutta (tapan@jcbose.ac.in) with details of the error and the corrections to be made with adequate reference and accession number of the corresponding information in other database(es), if possible. Please do mention the RHObase ID in your e-mail.
Go to top
How current is the information in RHObase?
The contents of RHObase presently include RHO entries (most of which are experimentally characterized) published in NCBI till December 2012. Information added thereafter will be included in the database very soon.
Go to top
How often is the database updated?
This is the first version of the database. It is expected to be updated every six months.
Go to top
How can I submit a new entry to the RHObase?
You may email Dr. Tapan K. Dutta (tapan@jcbose.ac.in) or Dr. Sudipto Saha (ssaha4@jcbose.ac.in) with any new entries that you would like to see included. The new entry will be added to the database only after verification.
Go to top
Can I download the database?
You may download conents of the database (sequences in Fasta format and substrate structures in SDF MOL file formats) from the "DOWNLOADS" page
Go to top
How do I make suggestions or request a new feature be added to the RHObase?
We are in a process of adding new features to the database. We encourage users to send back their feedback and suggestions so as to improve the content and features of RHObase. Please email Dr. Tapan K. Dutta (tapan@jcbose.ac.in) with any new feature(s) or query that you would like to see included, with as much detail as possible.
Go to top
Are there any suggested readings related to the contents of this database?
You may refer to the following publications to know more about RHOs and other related databases:
1) Gibson DT, and Subramanian V (1984). Microbial degradation of aromatic hydrocarbons. In "Microbial degradation of organic compounds". (ed. D.T. Gibson). New York: Marcel Dekker, Inc. pp. 181-252.
2) Boyd DR and Sheldrake GN (1998). The dioxygenase-catalysed formation of vicinal cis-diols. Nat. Prod. Rep. 15:309-3224.
3) Hudlicky T, Gonzalez D and Gibson DT (1999). Enzymatic dihydroxylation of aromatics in enantioselective synthesis: Expanding asymmetric methodology. Aldrichimica Acta. 32:35-62.
4) Gibson DT and Parales RE (2000). Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243.
5) Arora PK, Kumar M, Chauhan A, Raghava GPS and Jain RK (2009). OxDBase: a database of oxygenases involved in biodegradation. BMC Res. Notes. 2:67.
6) Gao J, Ellis LBM and Wackett LP (2010). The University of Minnesota Biocatalysis/Biodegradation Database: improving public access. Nucl. Acid. Res. 38:D488-D491.
7) Parales RE and Resnick SM (2010). Applications of aromatic hydrocarbon dioxygenases. In "Biocatalysis in the Pharmaceutical and Biotechnology Industries" (ed. R.N. Patel), CRC Press. pp. 299-317.
8) Mallick S, Chakraborty J, and Dutta TK (2011). Role of oxygenases in guiding diverse metabolic pathways in the bacterial degradation of low-molecular weight polycyclic aromatic hydrocarbons: A review. Crit. Rev. Microbiol. 37:64-90.
9) Chakraborty J, Ghosal D, Dutta A and Dutta TK (2012). An insight into the origin and functional evolution of bacterial aromatic ring-hydroxylating oxygenases. J. Biomol. Struct. Dyn. 30:419-36.
Go to top
| 


