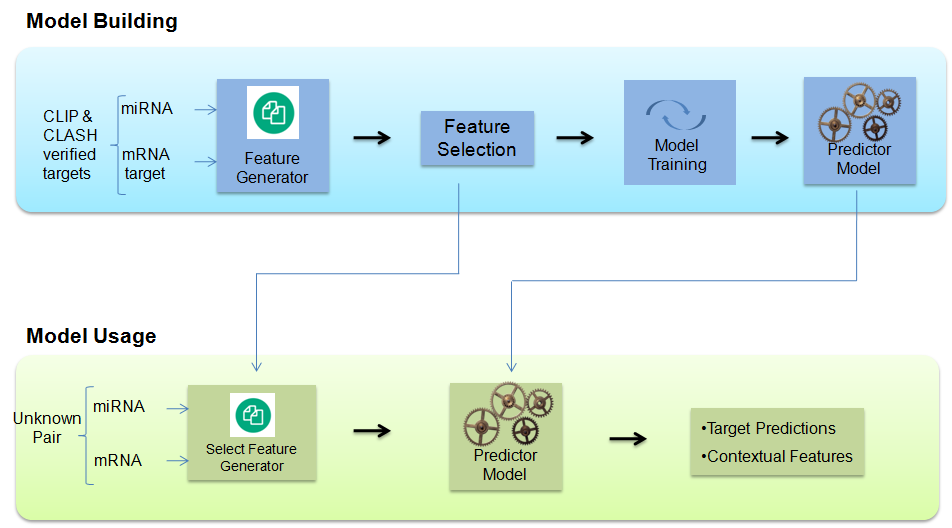
Overall Framework
Model Building
miRTPred model is built by using experimentally validated miRNA-mRNA interactions from CLIP and CLASH. Thereafter, context features of the target binding site are generated from which the enriched features are selected. Finally several ensemble and non ensemble based models are trained, to select the optimal blended ensemble based predictor model with maximum predictive performance.
Model Usage
The miRNA-mRNA pair given by the user is used to generate the context features. These features (generated in the previous step) are used by the predictor model to finally generate the predictions
Standalone Execution Workflow
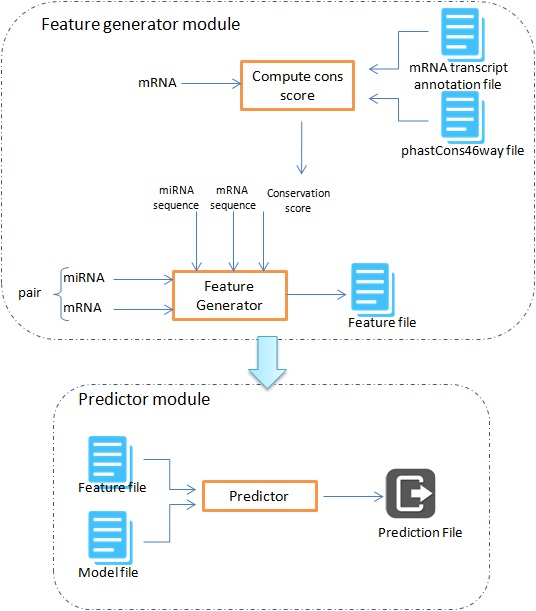
Feature Generator Module
This module of the miRTPred's execution workflow, first computes the phastCons conservation score using phastcons46way conservation score from UCSC. The feature generator program then uses this conservation file, miRNA and mRNA reference sequence files to generate the context feature file for the user's miRNA and mRNA pair of interest.
Predictor Module
The feature file and the optimal predictor model file are then used to predict the target binding site and associate a p-score. The p-score gives the probability of miRNA-mRNA interaction at that binding site.
