
 |
|---|
PVT (Pipelined Version of TopHat) is an efficient spliced alignment tool for RNA-Seq reads. It is capable of aligning both single-end and paired-end RNA-Seq reads to mammalian-sized genomes and finding out both annotated and novel splice junctions. In PVT , we take up a modular approach by breaking TopHat’s serial execution into a pipeline of multiple stages, thereby increasing the degree of parallelization and computational resource utilization. The workflow for both single end and paired end reads is so designed that increases its efficiency with respect to its computation time and computational resource utilization (in terms of CPU and memory utilization). Single End Read Analysis: In PVT for single end reads, we parallelized the steps where the computational resource is under utilized and removed the redundant steps during the execution of each dataset which improved its efficiency and enforced utilization of computational resource along with the reduction of the execution time. Paired End Read Analysis: For paired end reads we rescheduled the execution of each steps and distributed the job in separate machines, in addition to removing the redundant steps during the execution of each dataset. |
||||
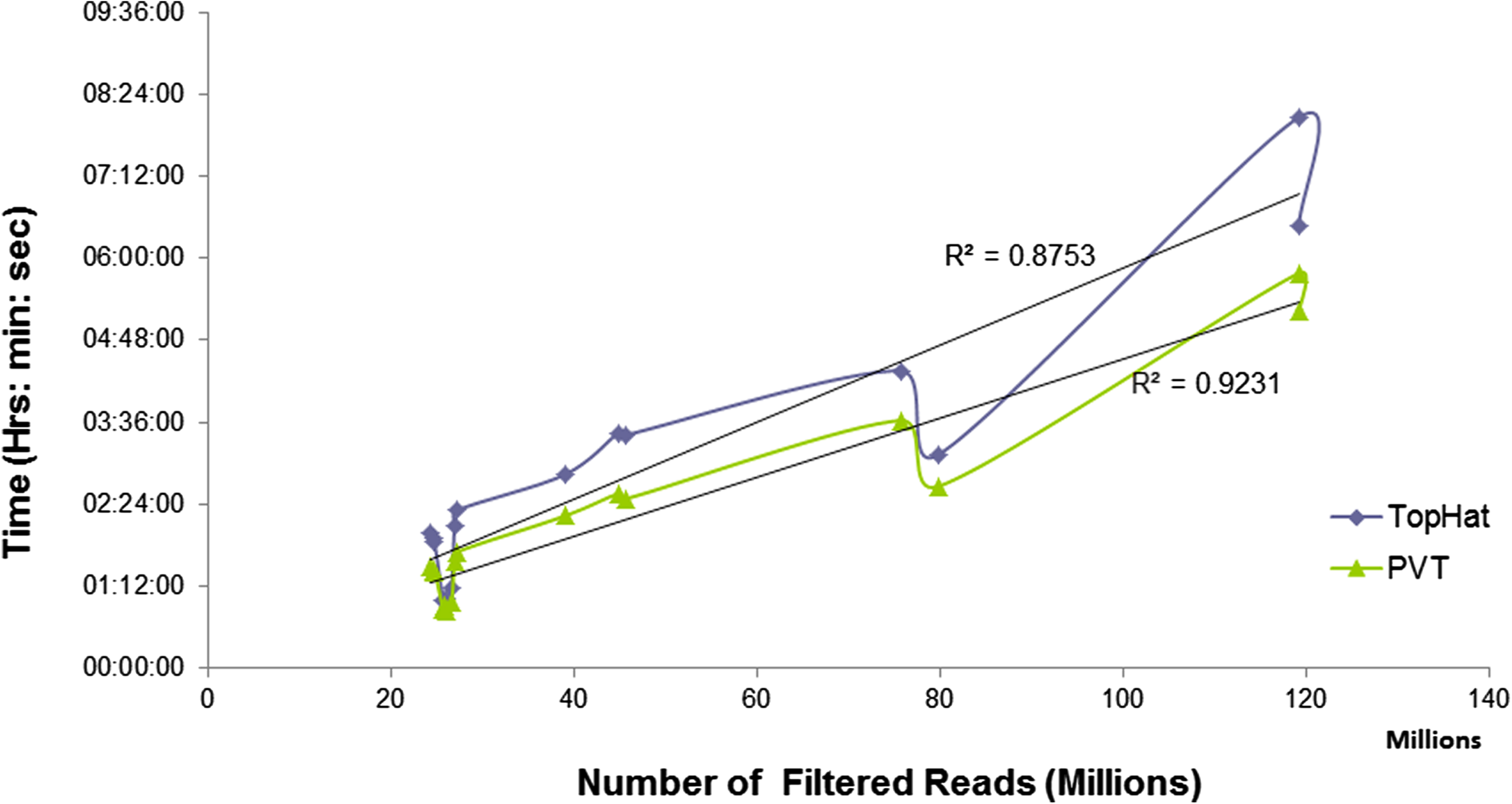 |
| Figure: Comparison of the improvement of PVT execution time with that of TopHat for single end reads. Each of the curves is fitted with a linear regression line. |
| Note: If you find PVT (Pipelined Version of TopHat) useful, please cite us at: "PVT: An Efficient Computational Procedure to Speed up Next-generation Sequence Analysis" BMC bioinformatics 15.1 (2014): 167. PMID: 24894600 |