|
What is PPIMpred?
Prediction of Protein Protein Interaction Modulators abbreviated as "PPIMpred" is a SVM based webserver for high throughput screening of small molecules targeting Protein- Protein Interactions (PPIs).It allows high throughput screening of small molecules for targeting PPIs using the physicochemical properties of known orthosteric PPIMs.
Go to top
What are PPIMs?
Protein - protein interactions (PPIs) are vital to a cellular process and disruption of an important PPI involved in disease process may open up a new drug discovery. The disruption of the PPI interface can be done using small chemicals known as PPI modulators (PPIMs). A few of known small chemical PPIMs such as Nutlin-3a, ABT-263 and GX15-070 against these three PPIs were clinically tested.
Go to top
Why do we focus on PPIM?
screening of small molecules for targeting PPIs using the physicochemical properties of known orthosteric PPIMs. PPIMpred is based on three important PPIs, namely Mdm2/P53, Bcl2/Bak and c-Myc/Max.These three transient interactions play roles in cell growth or programmed cell death (apoptosis) that indicates their involvement in different stages of cancer. Mdm2 is the cellular inhibitor of P53, a tumor suppressor. c-Myc/Max complex is a transcriptional activator, whereas Bcl2/Bak plays central role in mitochondrion-mediated apoptotic cell death.
Go to top
How users will give inputs to the PPIMpred?
User should fill all information in all the fields such as 10 physico-chemical properities (Molecular Weight,Molecular Formula,XLogP3,Hydrogen Bond Donor Count,Hydrogen Bond Acceptor Count,Rotatable Bond Count,Topological Polar Surface Area,Heavy Atom Count,Complexity,Defined Atom Stereocenter Count,Defined Bond Stereocenter Count,Covalently-Bonded Unit Count), target selection ( Mdm2/P53,c-MYC/MAX & BCL2/BAK) and choose threshold value . One example was given on the home page. No field may not be left blank. In the batch option users may input as the example shown in the page.
Single Input: 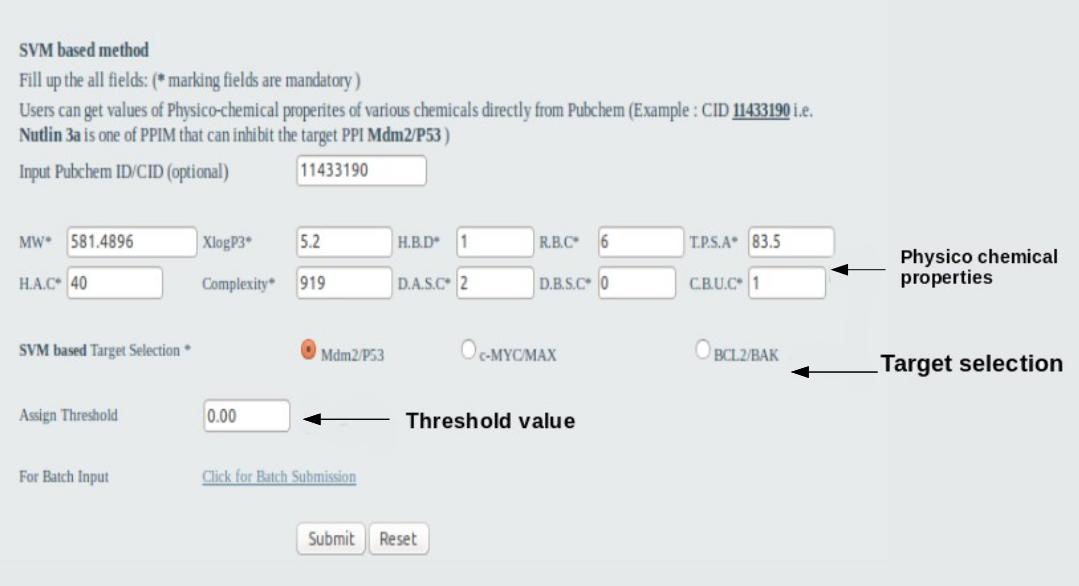 Batch input:
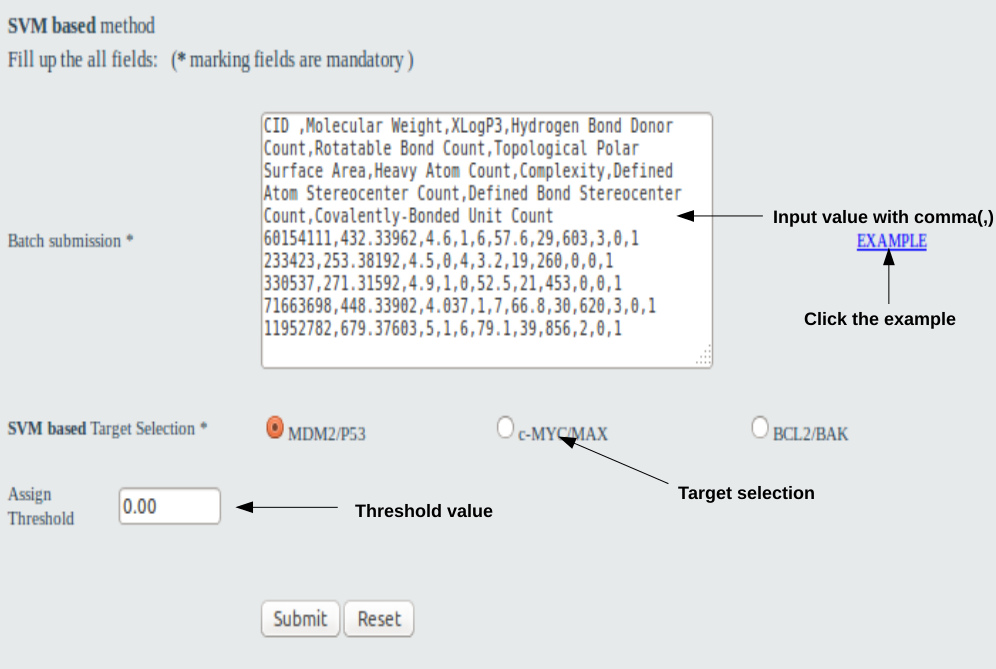
Go to top
How dataset will be prepared for singe input/batch input?
There are 7 steps for preparing dataset for singe input/batch inputs. such as:
- Step 1: First users may visit to the Pubchem physico-chemical properties section and Copy.
- Step 2: Users may paste Copied section directly to the Excel sheet.
- Step 3: Then users paste again by Paste special with transpose.
- Step 4: After that delete selected colored section such as Molecular Formula, Hydrogen Bond Acceptor Count, Exact Mass, Monoisotopic Mass, Formal Charge, Isotope Atom Count, Undefined Atom Stereocenter Count & Undefined Bond Stereocenter Count.
- Step 5: Rest of the sheet remains 10 selected descriptors/properties like as Molecular Weight, XLogP3, Hydrogen Bond Donor Count,Rotatable Bond Count,Topological Polar Surface Area, Heavy Atom Count,Complexity,Defined Atom Stereocenter Count, Defined Bond Stereocenter Count & Covalently-Bonded Unit Count with values (Remove also symbols units). Then users also may add one more column named Chemical Id/CID.
- Step 6: The process will be repeated if more than one inputs. then save file .csv format and then copy all and insert the blank input area of batch input option. Please read the example file of inputs.
- Step 7: Finally the file will ready for submitting to PPIMpred.
Please see the example of input pattern before submit
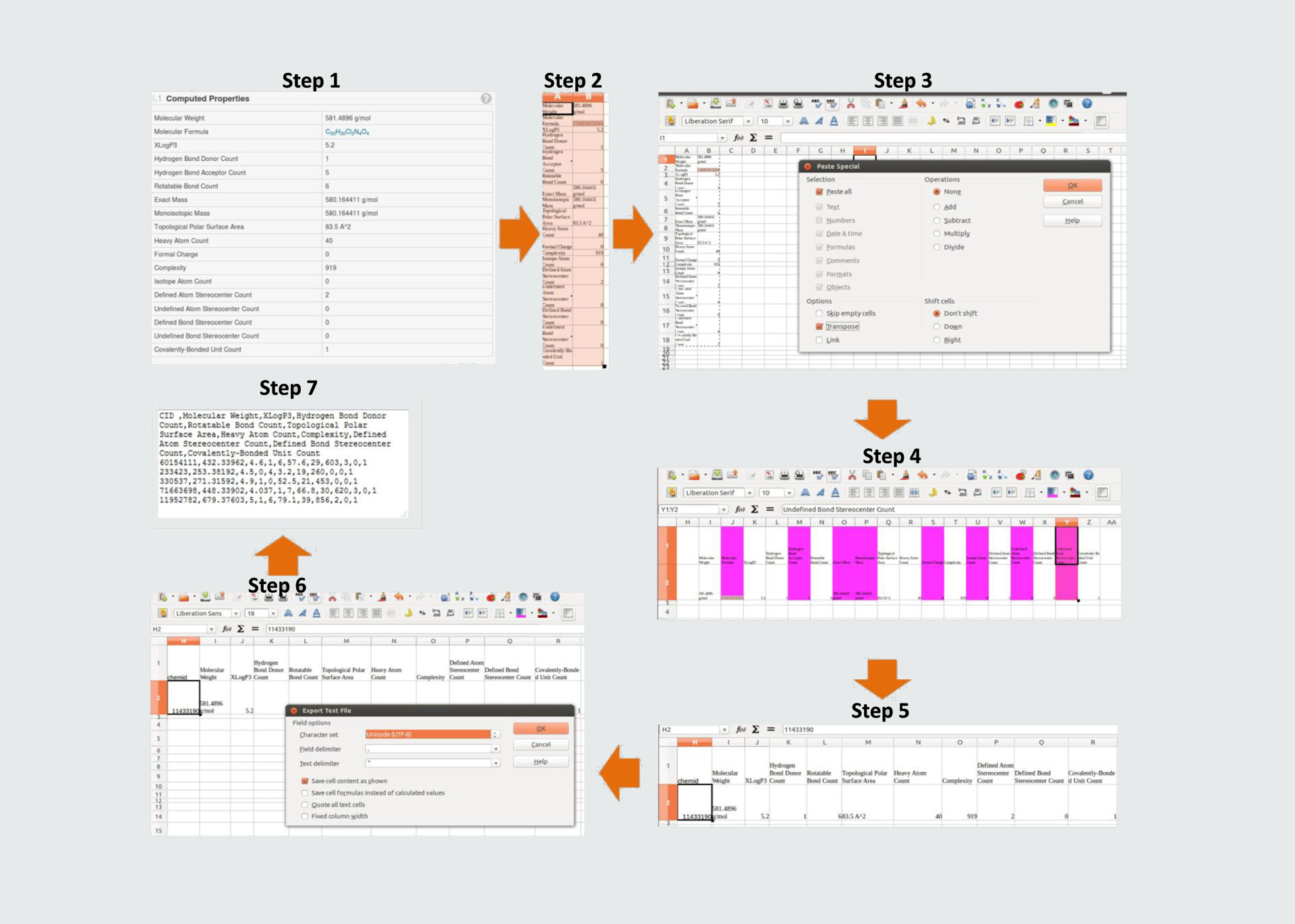
Go to top
how output of prediction result will interpret to the users?
There three major section of output page of prediction result such as prediction result section, Tabular result section and graphical section.
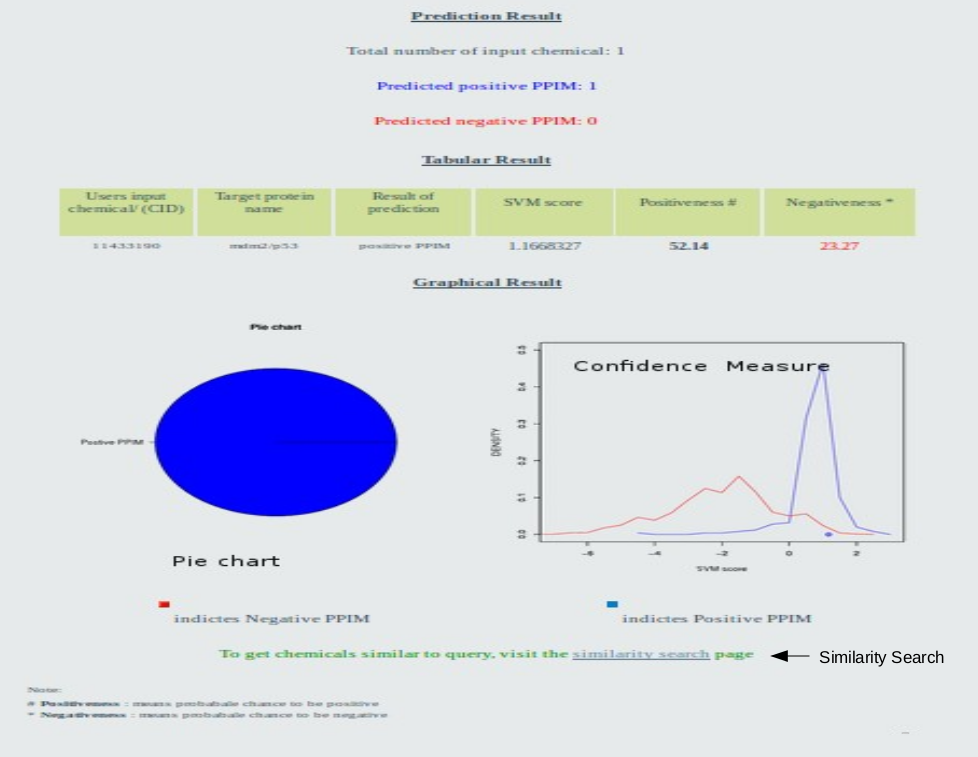
Go to top
Beside Prediction what is another application of PPIMpred web-server?
Beside classification (Yes/No), PPIMpred allow users to get the structural and chemical similarities with known modulators based on similarity search algorithm (Tanimoto Co-efficient) from “Similarity Search” page.
Go to top
What will be inputs of Similarity Search?
Users may draw chemical strcture JSME editor or paste MOL file of the input chemical directly to the blank text area. Then users may choose one of three the target and click the 'submit' button.
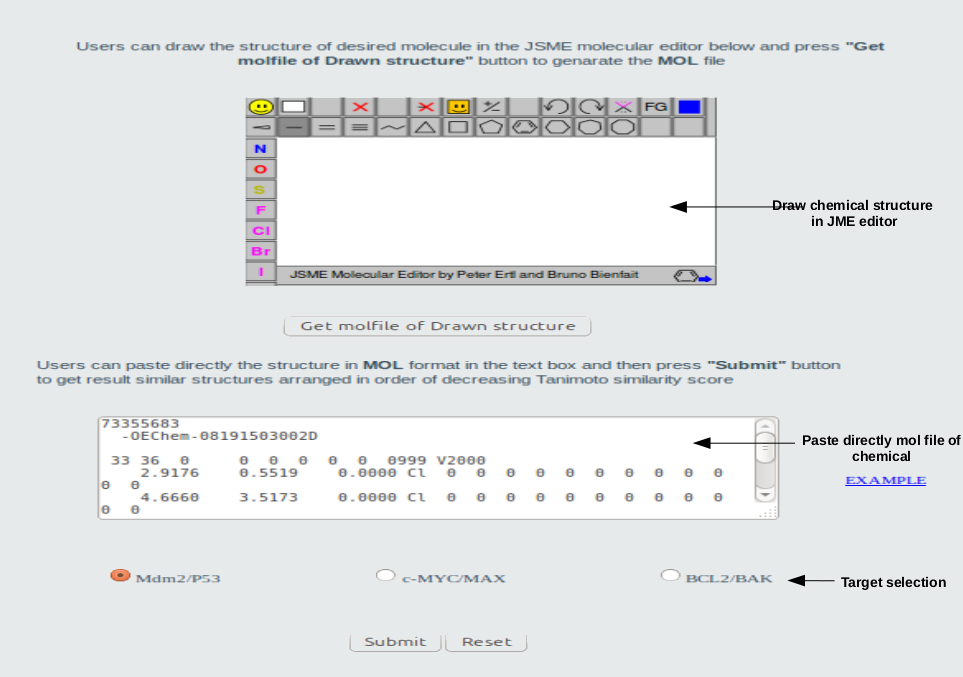
Go to top
how output of similarity search will interpret to the users?
After clicking the submit button, Users can show List of compounds similar to the query structure, arranged in order of decreasing Tanimoto similarity score (Click over the Pubchem/CID to know more about the chemical in the Pubchem):
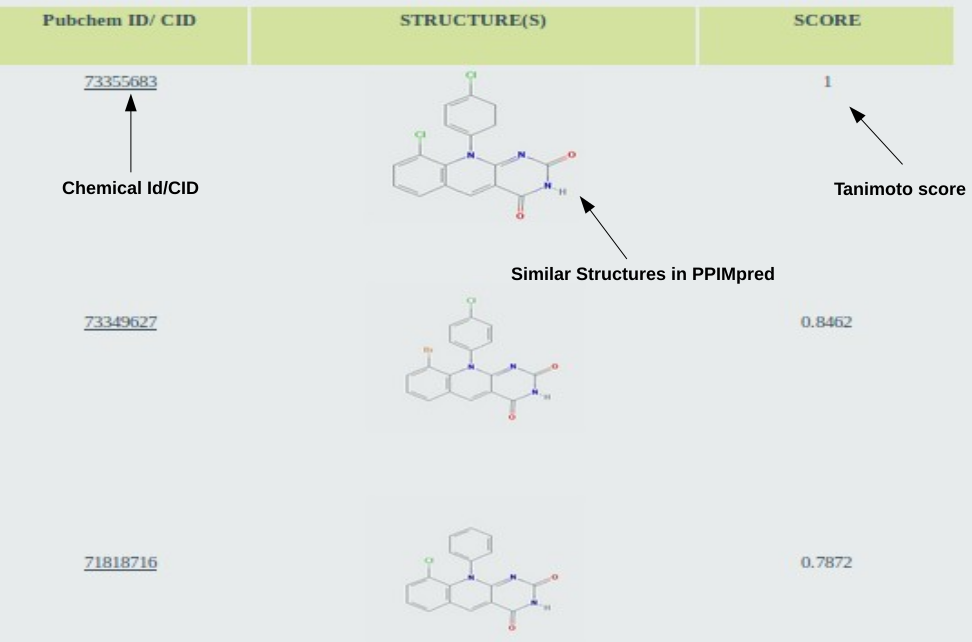
Go to top
Can user download the results and datasets?
Users may download all datasets and result in the PPIMpred webserver.
Go to top
What will be ouput of known PPIMs which were already clinically tested?
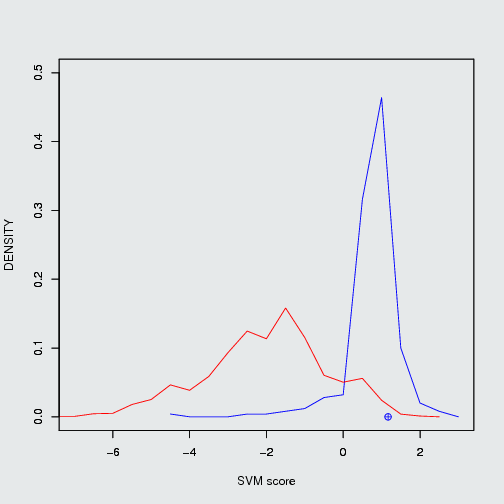
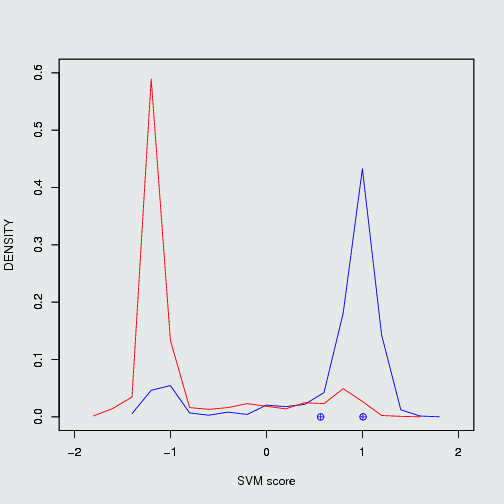
Nutlin-3a(Mdm2/P53), ABT-263(Bcl2/Bak) and GX15-070 (Bcl2/Bak) are known PPIMs which were clinically tested. The three PPIMs were evaluted by our SVM based algorithm, the output of three PPIMs are given below.
| Users input chemical/ (CID) |
Target protein-protein interaction complex |
Result of prediction |
SVM score |
Positiveness |
Negativeness |
| Nutlin 3a (11433190) |
Mdm2/P53 |
positive PPIM |
1.17 |
52.14% |
23.27% |
| ABT-263 |
Bcl2/Bak |
positive PPIM |
1.01 |
42.86 |
3.46 |
| GX15-070 |
Bcl2/Bak |
positive PPIM |
0.56 |
17.02% |
13.27% |
 |
 |
| Position of Nutlin 3a in frequency density plot of Mdm2/P53. |
Position of ABT-263 & GX15-070 in frequency density plot of Bcl2/Bak |
Are there any suggested readings/query related to the contents of this PPIMpred?
For queries and suggestion please contact Dr. Sudipto Saha (ssaha4@jcbose.ac.in) (ssaha4@gmail.com)
Go to top
Is there any Supplementary tables and figures for PPIMpred?
Any one can visit figshare website for supplementary tables and figures. click to visit
OR
Here PDF copy for supplementary tables and figures of PPIMpred is provided. Click to download
Cite this article:
Jana T, Ghosh A, Das Mandal S, Banerjee R, Saha S. 2017 PPIMpred:a web server for high-throughput screening of small molecules targeting protein–protein interaction.R. Soc. open sci.4: 160501. http://dx.doi.org/10.1098/rsos.1605
Go to top
|









