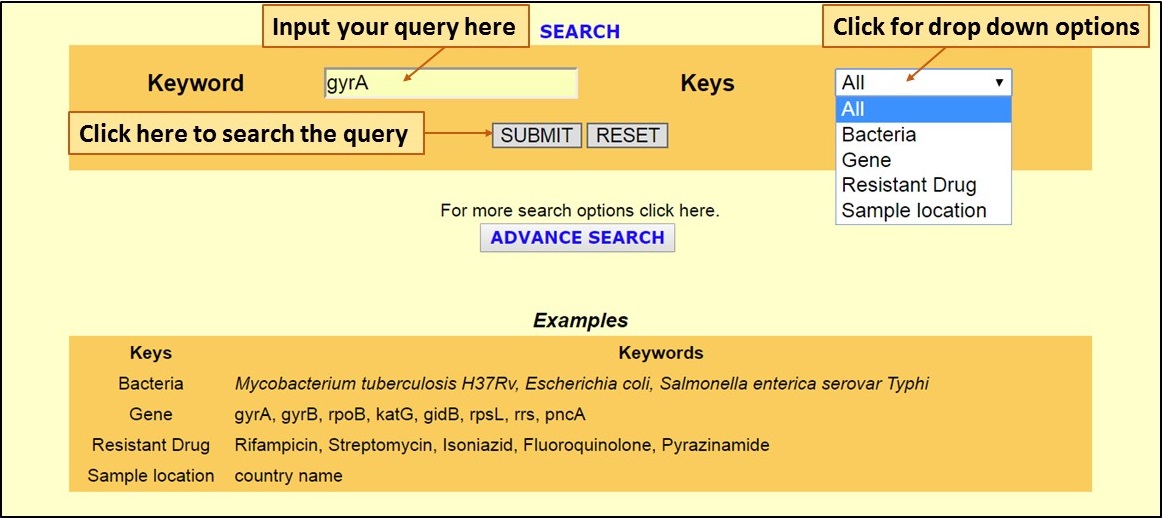
| [A] The Search option on the homepage allows users to query the database with specific keywords for Bacteria, Gene, Drug and Sample Location: |
 |
| The result page provides information on the genes associated with specific drug resistance in bacteria, nucleotide positions with mutation, aminoacid positions with mutations, PROVEAN score that indicates the functional impact of the mutation, sample location and links to the PubMed page of the referenced research articles that report the mutations. |
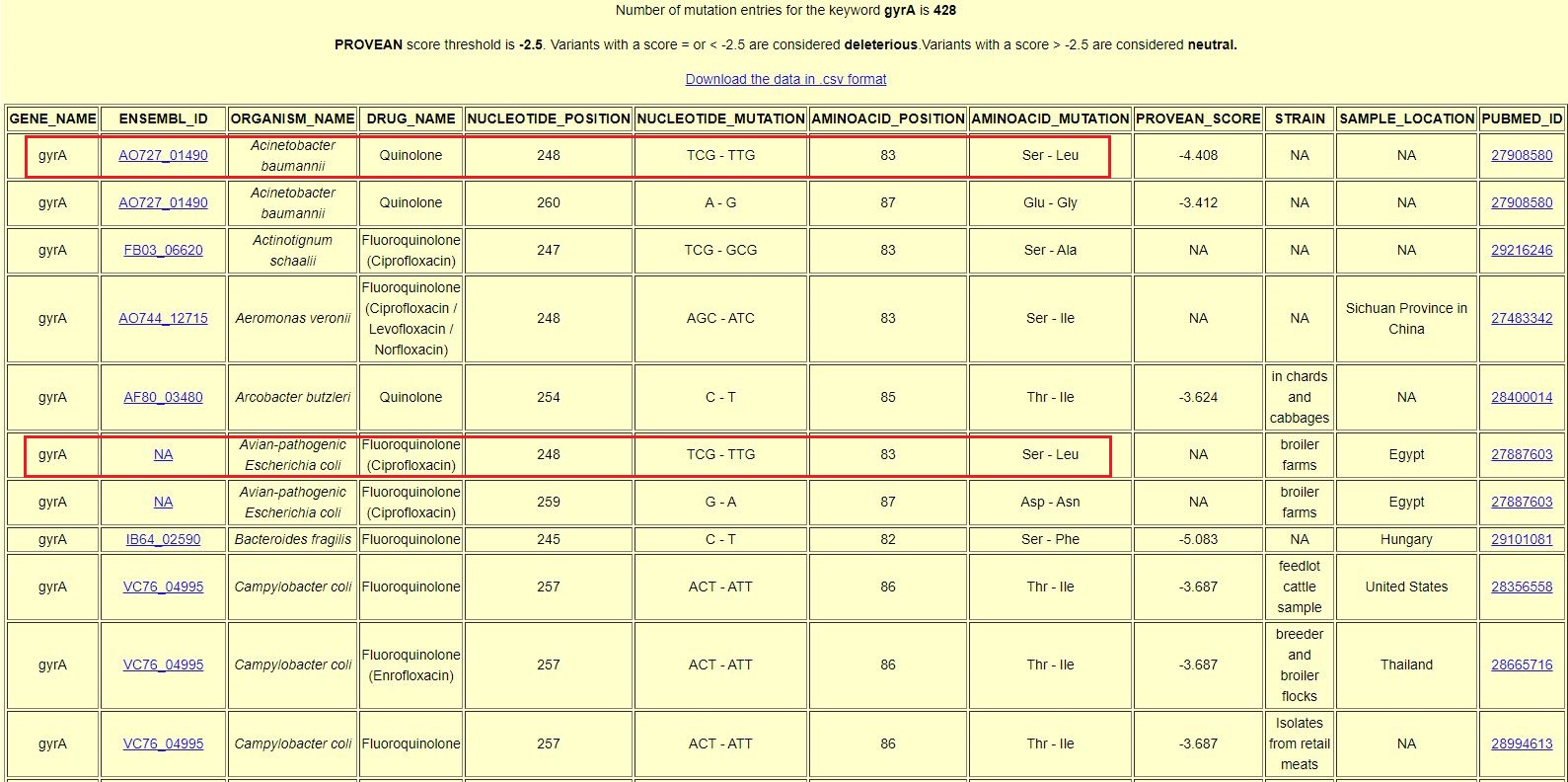 |
| The result for search query "gyrA" when "All" key is selected shows different bacteria (Coloured boxes) with mutations in gyrA related to Fluoroquinolone resistance. This phenomenon of same drug resistance (phenotype) in response to similar mutations (at nucleotide position 248 in Acinetobacter baumannii and Escherichia coli) in same gene (genotypic changes) is known as Homoplasy.
|
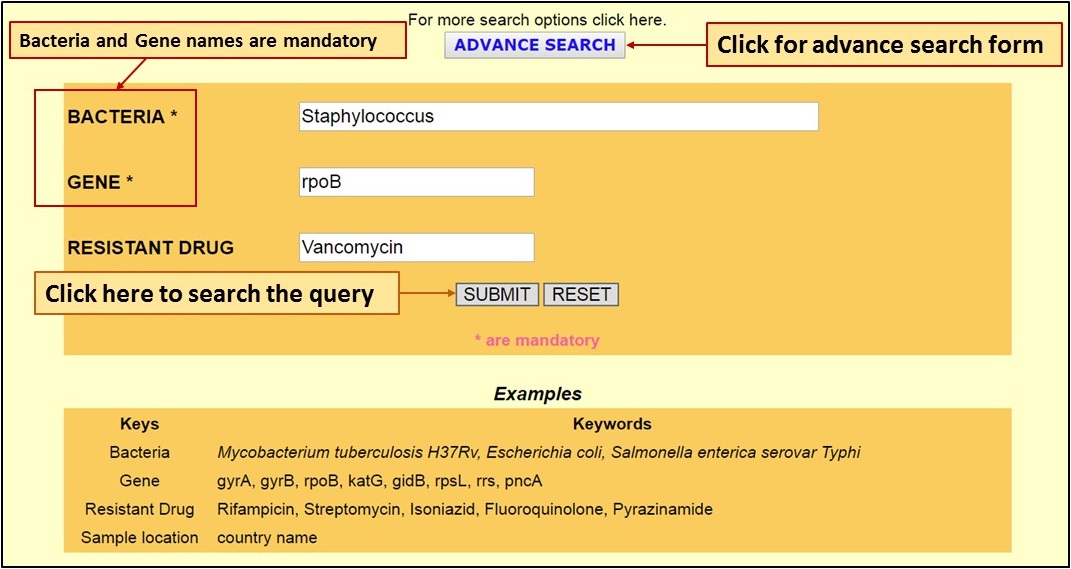
| [B] The Advance Search option on the homepage allows users to query the database with mandatory Bacteria and Gene name and optional Drug name: |
 |
| The result page of Advance search: |
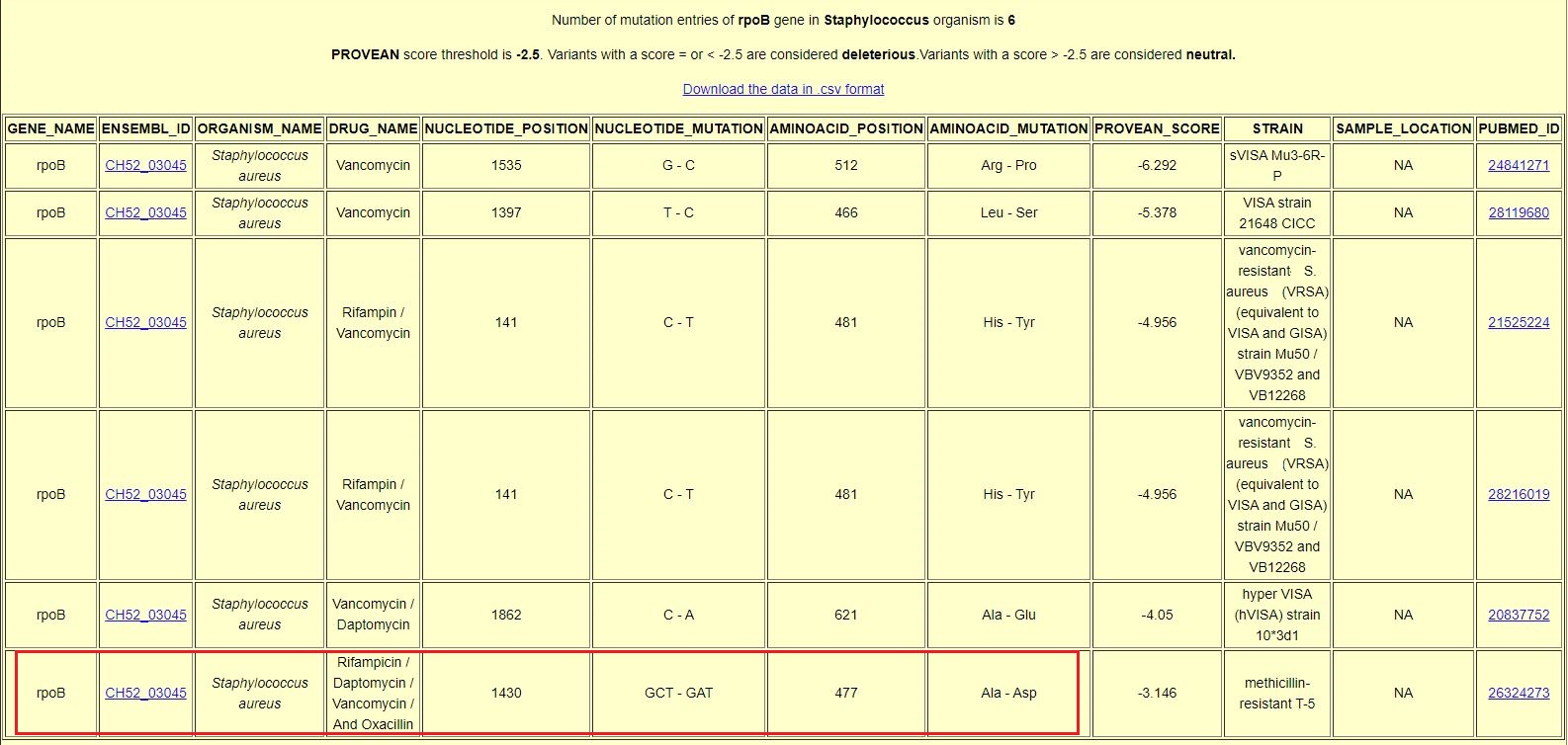 |
| The result for Advance search query where bacteria is "Staphylococcus", gene is "rpoB" and drug is "Vancomycin" shows an isolate of Staphylococcus aureus with a single mutation in rpoB associated with drug resistance to Rifampicin, Daptomycin, Vancomycin & Oxacillin (Coloured box). The phenomenon when a single mutation (genotypic change) at nucleotide position 1430 leads to multiple drug resistances (phenotypes) is known as Pleiotropy.
|
| [C] The Utility Page allows users to Blast Search the specific short nucleotide sequence stretches of drug resistance determining regions (DRDR) with reported mutations from DRAGdb: |
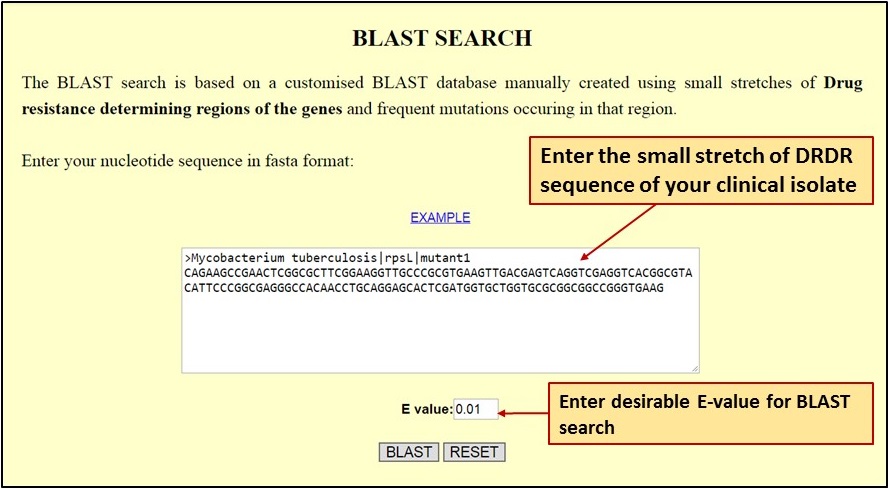 |
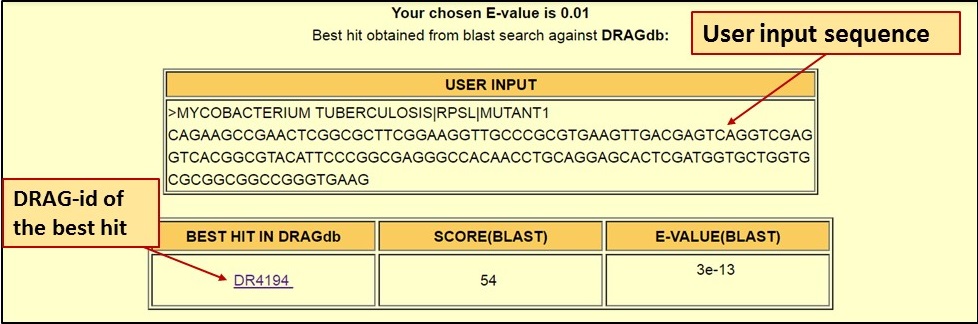
| The Blast Search result page shows the best hit, its blast score and E-value. The DRAGdb id of the best hit is given and it links to the entry information. |
 |
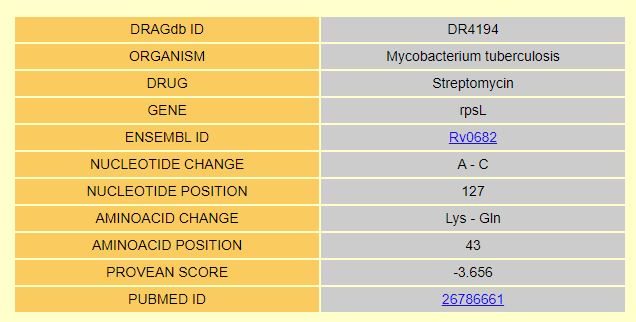
| The DRAG-id links to the entry page where all the information of the best hit is given. |
 |



