
| About lncRNA subtypes and ID assignment | |
|---|---|
1. lncRNA subtypes | |
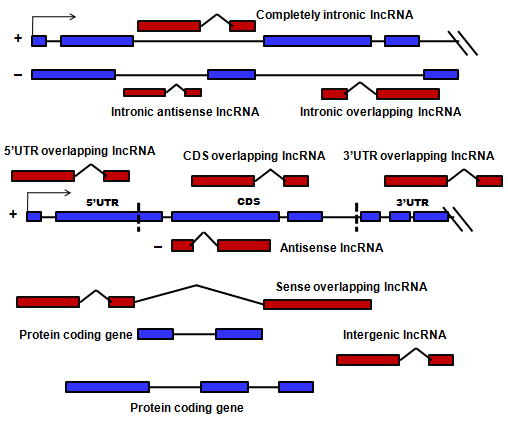 |
LncRBase hosts fourteen distinct subtypes of lncRNAs including our newly classified types and other transcript biotypes. These are as follows:
(1) 3UO, 3/UTR overlapping lncRNAs overlapping any 3/UTR exon in the sense strand. (2) 5UO, 5/UTR overlapping lncRNAs overlapping any 5/UTR exon in the sense strand lncRNA (3) CDS, CDS overlapping lncRNAs overlapping any protein coding CDS exon. (4) LI, Intergenic (linc) lncRNAs transcribed from in between two gene loci (5) AN, Antisense lncRNAs intersecting any exon of a protein-coding locus on the opposite strand (6) CI, Completely Intronic transcripts reside within introns of a coding gene, but do not intersect any exons. (7) IA, Intronic Antisense lncRNAs that completely overlap with an intron in the opposite strand (8) IO, Intron Overlapping lncRNA splice variants of a gene, contain intronic sequence (9) PS, Pseudogene transcripts having homology to protein coding transcripts but containing disrupted coding sequence. (10) SO, Sense Overlapping lncRNAs containing a coding gene in its intron on the same strand (11) AO, Ambiguous ORF transcripts believed to be protein coding, but with more than one possible open reading frame (12) PT, Processed Transcript not containing an ORF and cannot be placed in any of the above categories (13) MI, miscRNA from the Ensembl transcript dataset. (14) NC, Non coding transcripts not falling in any of the above mentioned categories. |
2. Assignment of lncRNA IDs
Based on the chromosomal location, we assigned unique indentifiers to each transcript. Human lncRNA transcript identifiers (ID) begin with hsaLB. Human lncRNA transcripts have been designated a unique identifier as hsaLB_[type]_[numeral]. 'type' is the distinct subtype of the lncRNA transcript IDs. The numeral ({primary}.{secondary}) is assigned as per the chromosomal location of the transcript. Multiple transcripts from the same genomic locus have same {primary} number with increasing {secondary} number which represents different cDNA variants from the locus.
Similar naming convention has been followed for mouse lncRNA transcripts, preceded by 'mmu' prefix. |
| Search LncRBase | |
|---|---|
1. LncRNA transcripts | |
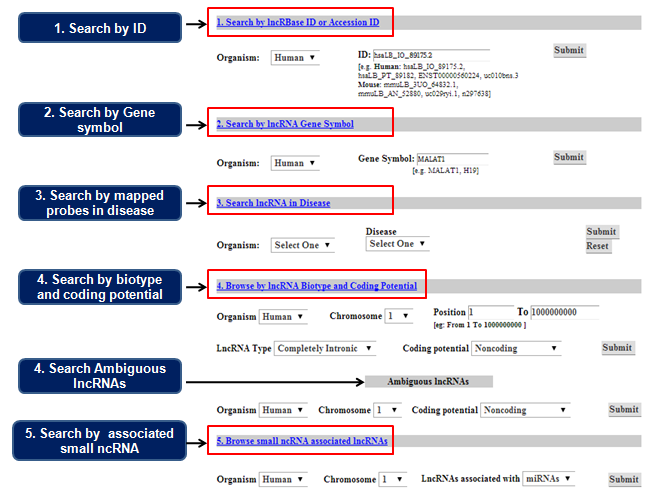 |
4 search options are available in LncRBase. 1. Search by Accession ID. Type a. LncRBase ID to view detailed information on that transcript. or b. Original ID (Ensembl/Noncode v3.0/UCSC Gene ID/Body Map lincRNA/H-InvDB 8.0 ). First you will be shown the number of entries of that particular ID in LncRBase. Clicking on a particular entry displays detailed information about that lncRNA. 2. Search by Gene Symbol. To search for a specific lncRNA, type its gene symbol(HGNC/MGI). You will be shown the number of entries for that gene in LncRBase and associated literature. Clicking on the particlar gene loci will show you the details of the gene and correponding transcript variants that are hosted in LncRBase. 3. Search lncRNA in disease. Select a specific disease system to view which Affymerix probes (HG U133 Plus2 for human and 430.2 for mouse) are expressed in that disease.The probes are mapped to respective lncRNAs. 4. Browse by lncRNA biotype and/or chromosomal position. Select organism first.Then select any specific lncRNA biotype and/or chromosomal position to view the specific lncRNAs in that region.You can further filter your query by selecting type of lncRNA you want to search, based on their coding potential, i.e. Noncoding or Putatively coding. A special feature is that you can search for Ambiguous lncRNAs also, that is those lncRNAs which have come under more than one biotype and are open to more than one interpretation. 5. Browse small ncRNA associated lncRNAs Select organism first.Then select any specific lncRNA biotype and chromosome to view lncRNA-associated miRNA or piRNAs |
2. LncRNA expression
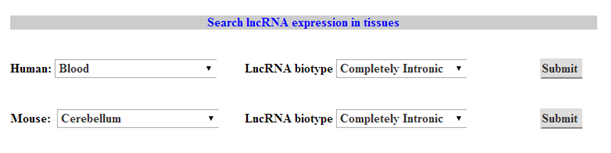
Select organiam and the particular tissue for which you want to check the expression of lncRNAs.You can filter your query by slecting for specific biotypes of lncRNA. in the output page you will be shown the total number of transcripts in that biotype and their average FPKM value in the selected tissue. |
View results of your search |
|
|---|---|
General Output | |
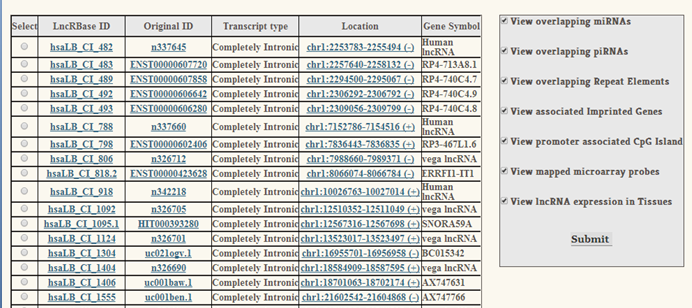 |
The first output page displays the general information of an lncRNA, like its LncRBase ID (assigned as per our genomic categorization protocol), Original ID (ID assigned by the database from where the transcript sequence was retrieved), biotype, genomic location, strand and gene symbol. A sidebar allows you to select extra attributes of the lncRNA of your interest. |
Detail Output
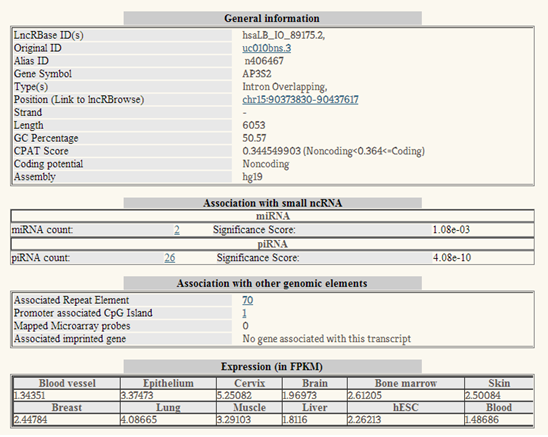
Clicking on an individual transcript ID in the general output page will direct you to a detailed output page. You can view LncRBase ID, Original ID, Alias ID , genomic location, transcript length,%GC,CPAT score,coding potential, associated miRNAs, associated piRNAs, number of microarray probes mapped to that lncRNA, associated disease, associated Imprinted gene, associated Repeat Element, lncRNA promoter information and lncRNA expression in various tissues.. |
lncRBrowse |
|
|---|---|
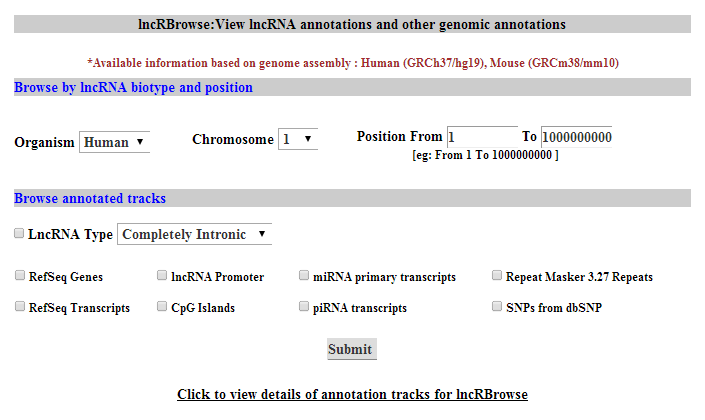 | |
The Browse option in the menubar takes you to the lncRBrowse page.You can select organism and then type specific chromosomal position.You can also view various annotated tracks:Refseq Genes, Refseq Transcripts, SNPs from dbSNP, Repeat Masker 3.27 Repeats, lncRNA biotype, lncRNA Promoter, CpG Island, miRBase premature miRNAs and piRNAs.You can select multiple tracks to view them together in the same genomic context. |
|
How to navigate through lncRBrowse | |
|
| |
|
| |
|
| |
|
| |
|
References:
| |
Click to view details of annotation tracks for lncRBrowse