ABOUT
Dataset:
There were some dataset used in EnPPIpred. Such as : i) Postive dataset, ii) Negative dataset I, iii) Negative dataset II, iv) Blind postive dataset & v) Blind negative dataset. The positive E. coli TAP-tag pull-down experimental protein-protein interactions (PPIs) data were downloaded from Bacteriome.org (Hu et al. TAP interaction dataset (AKA 'Core - experimental')). The negative E. coli protein pairs were generated by random pairing between proteins.
Methods:
Support Vector Machine (SVM) with 5-fold cross-validation technique was employed, to build a model for predicting protein-protein interactions (PPIs) in Enteropathogen. Different features including domain-domain associations (DDA), degree (No. of interacting partners present in positive protein-protein interactions network), amino acid composition (AAC) and dipeptide composition (DC) were tested in order to optimize the performance of proposed SVM model.
Table 1: SVM kernel-wise performance measures of E.coli dataset using 5-fold cross-validation technique. Optimal parameters(DDA, degree and amino acid composition (Default Hybrid)) and threshold were used for respective kernel.
| SVM Kernel | Sensitivity (%) | Specificity (%) | Accuracy (%) | PPV (%) | MCC | F1 Score (%) | ROC |
| Linear | 73 | 89 | 81 | 87 | 0.63 | 79.39 | 0.881 |
| Polynomial | 49 | 96 | 72 | 92 | 0.51 | 63.94 | 0.769 |
| Radial basis function | 77 | 86 | 82 | 85 | 0.64 | 80.80 | 0.878 |
| Sigmoid | 80 | 82 | 81 | 81 | 0.62 | 80.50 | 0.877 |
Figure 1: The ROC curves showing threshold independent performances of different kernels of SVM on E.coli dataset.
Table 2: SVM performance measures on different subsets of features in E. coli dataset, using RBF kernel. Optimal parameters were used for respec-tive subset of features.
| Features of Protein Pair Features | Vector Length | Sensitivity (%) | Specificity (%) | Accuracy (%) | PPV (%) | MCC | F1 Score (%) | Area under ROCcurve |
| Domain-Domain Association(DDA) | 1 | 32 | 92 | 62 | 80 | 0.57 | 45.71 | 0.633 |
| Degree | 1 | 83 | 79 | 81 | 80 | 0.62 | 81.47 | 0.874 |
| DDA and Degree | 2 | 80 | 82 | 81 | 81 | 0.62 | 80.50 | 0.874 |
| Amino acid composition (AAC) | 40 | 78 | 76 | 77 | 77 | 0.55 | 77.50 | 0.824 | DDA and AAC | 41 | 70 | 87 | 78 | 84 | 0.57 | 76.36 | 0.844 |
| Degree and AAC | 41 | 77 | 85 | 81 | 84 | 0.63 | 80.35 | 0.878 |
| DDA, Degree and AAC | 42 | 77 | 86 | 82 | 85 | 0.64 | 80.80 | 0.878 |
| Dipeptide Composition (DC) | 800 | 79 | 76 | 77 | 77 | 0.55 | 77.99 | 0.826 |
| Degree and DC | 801 | 83 | 79 | 81 | 80 | 0.62 | 81.47 | 0.868 |
| DDA, Degree and DC | 802 | 76 | 86 | 81 | 85 | 0.63 | 80.25 | 0.868 |
| DDA, AAC and DC | 841 | 71 | 88 | 79 | 85 | 0.60 | 77.37 | 0.849 |
| All Features (DDA, Degree,AAC and DC) | 842 | 76 | 87 | 82 | 85 | 0.63 | 80.25 | 0.882 |
Table 3: Different threshold wise SVM performance measures of E.coli dataset using 5-fold cross-validation, parameters t = 2 (RBF kernel), g = 0.001, c = 0.9, j = 1 and best subset of features (DDA, Degree and AAC)
| Threshold | Sensitivity(%) | Specificity(%) | Accuracy(%) | PPV(%) | MCC | F1 Score(%) |
| 2.00 | 0 | 20 | 10 | 20 | 0.01 | 0.00 |
| 1.90 | 0 | 40 | 20 | 30 | 0.01 | 0.00 |
| 1.80 | 0 | 80 | 40 | 73 | 0.02 | 0.00 |
| 1.70 | 0 | 80 | 40 | 63 | 0.02 | 0.00 |
| 1.60 | 1 | 100 | 50 | 84 | 0.04 | 1.98 |
| 1.50 | 2 | 100 | 51 | 90 | 0.08 | 3.91 |
| 1.40 | 3 | 100 | 51 | 91 | 0.11 | 5.81 |
| 1.30 | 6 | 99 | 53 | 91 | 0.16 | 11.26 |
| 1.20 | 13 | 99 | 56 | 93 | 0.24 | 22.81 |
| 1.10 | 28 | 98 | 63 | 93 | 0.36 | 43.04 |
| 1.00 | 47 | 97 | 72 | 93 | 0.50 | 62.44 |
| 0.90 | 57 | 95 | 76 | 92 | 0.57 | 70.39 |
| 0.80 | 62 | 94 | 78 | 91 | 0.59 | 73.75 |
| 0.70 | 66 | 93 | 79 | 90 | 0.61 | 76.15 |
| 0.60 | 69 | 91 | 80 | 89 | 0.62 | 77.73 |
| 0.50 | 71 | 90 | 81 | 88 | 0.62 | 78.59 |
| 0.40 | 73 | 89 | 81 | 87 | 0.63 | 79.39 |
| 0.30 | 74 | 88 | 81 | 87 | 0.63 | 79.98 |
| 0.20 | 75 | 88 | 81 | 86 | 0.63 | 80.12 |
| 0.10 | 76 | 87 | 82 | 85 | 0.64 | 80.25 |
| 0.00 | 77 | 86 | 82 | 85 | 0.64 | 80.80 |
| -0.10 | 78 | 85 | 81 | 84 | 0.63 | 80.89 |
| -0.20 | 79 | 84 | 81 | 83 | 0.63 | 80.95 |
| -0.30 | 80 | 83 | 81 | 82 | 0.62 | 80.99 |
| -0.40 | 81 | 82 | 81 | 82 | 0.63 | 81.50 |
| -0.50 | 82 | 81 | 81 | 81 | 0.62 | 81.50 |
| -0.60 | 83 | 79 | 81 | 80 | 0.62 | 81.47 |
| -0.70 | 84 | 78 | 81 | 79 | 0.62 | 81.42 |
| -0.80 | 86 | 75 | 81 | 78 | 0.61 | 81.80 |
| -0.90 | 88 | 72 | 80 | 76 | 0.60 | 81.56 |
| -1.00 | 91 | 61 | 76 | 70 | 0.55 | 79.13 |
| -1.01 | 92 | 58 | 75 | 69 | 0.53 | 78.86 |
| -1.02 | 92 | 55 | 74 | 67 | 0.51 | 77.53 |
| -1.03 | 92 | 51 | 72 | 66 | 0.48 | 76.86 |
| -1.04 | 93 | 48 | 71 | 64 | 0.46 | 75.82 |
| -1.05 | 94 | 44 | 69 | 63 | 0.43 | 75.44 |
| -1.06 | 95 | 39 | 67 | 61 | 0.40 | 74.29 |
| -1.07 | 95 | 34 | 65 | 59 | 0.37 | 72.79 |
| -1.08 | 96 | 30 | 63 | 58 | 0.34 | 72.31 |
| -1.09 | 97 | 26 | 61 | 56 | 0.32 | 71.01 |
| -1.10 | 97 | 21 | 59 | 55 | 0.28 | 70.20 |
| -1.20 | 99 | 3 | 51 | 51 | 0.09 | 67.32 |
| -1.30 | 100 | 1 | 50 | 50 | 0.06 | 66.67 |
| -1.40 | 100 | 0 | 50 | 50 | 0.03 | 66.67 |
| -1.50 | 100 | 0 | 50 | 50 | 0.01 | 66.67 |
| -1.60 | 100 | 0 | 50 | 50 | 0.00 | 66.67 |
| -1.70 | 100 | 0 | 50 | 50 | 0.00 | 66.67 |
| -1.80 | 100 | 0 | 50 | 50 | 0.00 | 66.67 |
| -1.90 | 100 | 0 | 50 | 50 | 0.00 | 66.67 |
| -2.00 | 100 | 0 | 50 | 50 | 0.00 | 66.67 |
Table 4: Performance measures on blind E. coli protein-protein interactions (PPIs) dataset (337 positive set was obtained from the Protein Data Bank (PDB) and 337 negative set was obtained from random pairs), using proposed optimal SVM model.
| Threshold | Sensitivity(%) | Specificity(%) | Accuracy(%) | PPV(%) | MCC | F1 Score(%) |
| 1.00 | 6 | 99 | 47 | 92 | 0.14 | 11.27 |
| 0.90 | 9 | 99 | 49 | 89 | 0.17 | 16.35 |
| 0.80 | 11 | 98 | 49 | 89 | 0.18 | 19.58 |
| 0.70 | 13 | 98 | 50 | 87 | 0.19 | 22.62 |
| 0.60 | 14 | 97 | 51 | 86 | 0.19 | 24.08 |
| 0.50 | 15 | 97 | 51 | 85 | 0.20 | 25.50 |
| 0.40 | 17 | 96 | 52 | 85 | 0.21 | 28.33 |
| 0.30 | 19 | 96 | 53 | 86 | 0.22 | 31.12 |
| 0.20 | 19 | 96 | 53 | 85 | 0.22 | 31.06 |
| 0.10 | 19 | 96 | 53 | 85 | 0.22 | 31.06 |
| 0.00 | 20 | 95 | 53 | 85 | 0.23 | 32.38 |
| -0.10 | 21 | 95 | 53 | 84 | 0.23 | 33.60 |
| -0.20 | 22 | 95 | 54 | 85 | 0.24 | 34.95 |
| -0.30 | 24 | 95 | 55 | 85 | 0.25 | 37.43 |
| -0.40 | 24 | 94 | 55 | 84 | 0.25 | 37.33 |
| -0.50 | 27 | 94 | 56 | 85 | 0.27 | 40.98 |
| -0.60 | 28 | 93 | 57 | 84 | 0.27 | 42.00 |
| -0.70 | 31 | 93 | 58 | 85 | 0.30 | 45.43 |
| -0.80 | 33 | 93 | 60 | 86 | 0.32 | 47.70 |
| -0.90 | 36 | 92 | 61 | 84 | 0.32 | 50.40 |
| -1.00 | 47 | 85 | 63 | 79 | 0.33 | 58.94 |
| -1.01 | 49 | 83 | 64 | 79 | 0.34 | 60.48 |
| -1.02 | 51 | 82 | 65 | 78 | 0.34 | 61.67 |
| -1.03 | 53 | 78 | 64 | 76 | 0.32 | 62.45 |
| -1.04 | 55 | 73 | 63 | 72 | 0.28 | 62.36 |
| -1.05 | 58 | 68 | 62 | 70 | 0.26 | 63.44 |
| -1.06 | 61 | 64 | 62 | 68 | 0.25 | 64.31 |
| -1.07 | 64 | 59 | 62 | 66 | 0.23 | 64.98 |
| -1.08 | 66 | 49 | 59 | 62 | 0.15 | 63.94 |
| -1.09 | 71 | 42 | 58 | 61 | 0.14 | 65.62 |
| -1.10 | 75 | 38 | 58 | 60 | 0.13 | 66.67 |
| -1.20 | 96 | 4 | 56 | 56 | 0.01 | 70.74 |
| -1.30 | 100 | 0 | 56 | 56 | 0.00 | 71.79 |
| -1.40 | 100 | 0 | 56 | 56 | 0.00 | 71.79 |
| -1.50 | 100 | 0 | 56 | 56 | 0.00 | 71.79 |
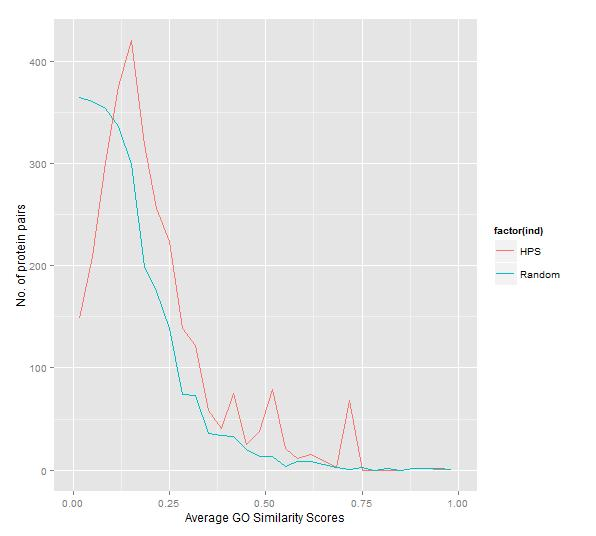
Fig 2: Frequency plot of average GO semantic similarity of HPS and Random S. Typhi PPIs.
Citations: Barman RK, Jana T, Das S, Saha S Prediction of Intra-species Protein-Protein Interactions in Enteropathogens Facilitating Systems Biology Study PLoS One. 2015 Dec 30;10(12):e0145648 PMID: 26717407