|
What is DAAB-V2? DAAB-V2 is an upgraded version of the Database of Allergy and Asthma Biomarkers. DAAB-V2 is a manually curated relational database enriched with four entity tables which are linked. In the first table (Master or Expression Table), besides, information on the expression profile of each entry, several new attributes (like Immunity Term, GO Molecular Function, GO Biological Process) have been incorporated. Each entry in the master table is hyperlinked to BioGRID ID for corresponding protein-protein interaction information. The other three newly incorporated tables contain information associated with Single Nucleotide Polymorphism, protein structures and drugs respectively.
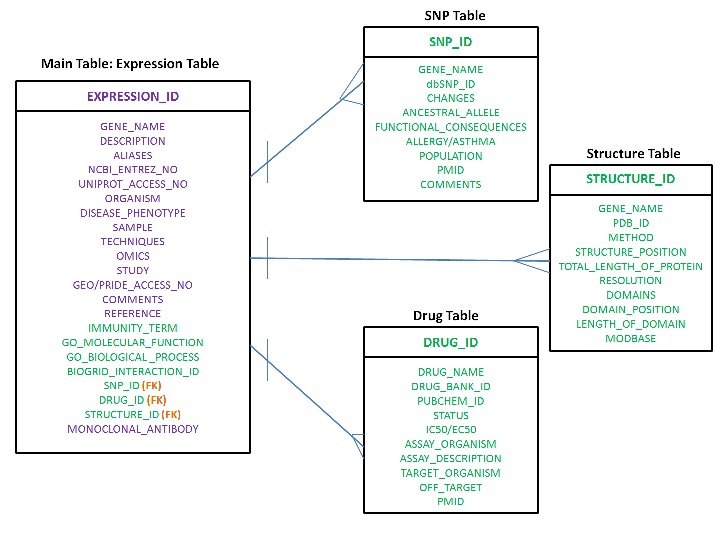
What is the Entity Relationship Diagram for the relational database DAAB-V2? The Entity Relationship Diagram (ER Diagram) for DAAB-V2 is as follows:
Figure: ER-diagram of DAAB (Version: 2).
The expression table serves as the master table with a primary key of EXPRESSION_ID. The master table is connected to three other tables namely, SNP Table, Structure Table and Drug Table using foreign keys SNP_ID, STRUCTURE_ID and DRUG_ID respectively. FK denotes the foreign keys in the master table. SNP_ID, STRUCTURE_ID and DRUG_ID serves as primary key for the SNP Table, Structure Table and Drug Table respectively. The purple coloured attributes denote those attributes that were already present in DAAB (Version 1) and the green coloured attributes have been newly incorporated in this newer version of DAAB-V2.
What are the new attributes included in DAAB-V2? The new attributes included in DAAB-V2 include the following:
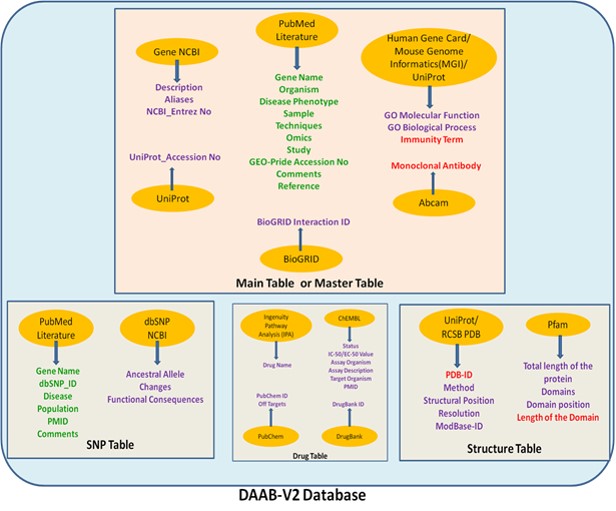
What are the different sources from which data has been collected for DAAB-V2? The differentially expressed bio-molecules in allergic diseases and asthma were manually curated from published literatures available in NCBI PubMed. These differentially expressed genes/proteins were further enriched manually with information on GO terms, structural details (including resolved structures and functional domains from RCSB PDB and pfam), available drugs and have been hyperlinked to BioGRID for relevant protein-protein interaction details. Role of SNPs of these bio-molecules in asthma and allergy was excavated from PubMed and compiled manually with other details (like dbSNP ID, variants or changes, ancestral allele, functional consequences) from dbSNP. The detailed methodology of data collection for DAAB-V2 has been schematically represented by the following figure:
Figure: Schematic representation of the DAAB-V2 architecture. A graphical illustration of the workflow and data source and contained in DAAB-V2. The data sources are contained in yellowish orange coloured oval shapes. Attributes mentioned in green colour have been manually curated, those in red colour have been partially curated/ derived and the attributes in purple colour have been derived from other secondary databases.
How many entries are there in DAAB-V2? DAAB-V2 contains a total of 14705 entries.
What are the new features included in DAAB-V2? DAAB-V2 has the following newly incorporated features: a) Keyword Search: The Keyword Search allows the user to search the database using all or any one of the keys from the drop down list for different keys (like Disease, Organism, Omics Approach, Tissue Type, Gene, GO Molecular Function, GO Biological Process, Structure). b) Advance Search: The Advance search option includes a Logical query interface with AND, OR and NOT operators and a drop-down list for available options, thus, making it user-friendly. The Advance Search option allows the user to execute comprehensive search using various combinations of the different keys available in the drop-down list, namely, Disease, Organism, Tissue Type, Omics Approach, Gene. The query is executed in a left associative manner. 2. Browse Page: The Browse page allows the user to browse the data contained in the database in the following ways:
How does the search output page look in DAAB-V2? The search output shall appear as follows:
The main table is linked to the tables for SNP, Structure and Drug using the SNP_ID, STRUCTURE_ID and DRUG_ID respectively. The newly incorporated attributes have been highlighted by red colour.
What are useful links in DAAB-V2? The useful links available in DAAB-V2 include:PubMed, BioGRID, NCBI, Gene Expression Omnibus (GEO), PDB, PubChem, Drug Bank.
|