Ghosh Lab
Professor at Department of Biological Sciences, Bose Institute, Kolkata

Professor at Department of Biological Sciences, Bose Institute, Kolkata
Research
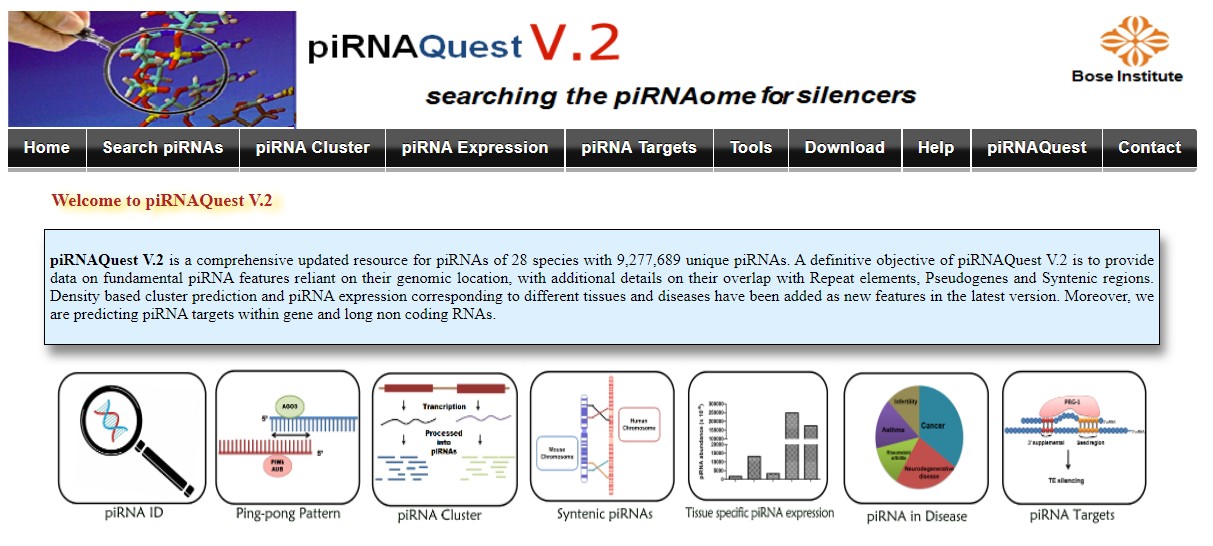
PIWI-interacting RNA (piRNA), a novel and emerging class of small noncoding RNA (sncRNA) ranges in length from 26-32 nucleotides. This sncRNA is a potent player in guiding the vital regulatory processes within a cellular system. To arrange classified information on piRNAs, we present piRNAQuest- a unified and comprehensive database on multispecies piRNAs. This database provides piRNA annotation based on their localization in gene, intron, intergenic, CDS, 5/UTR, 3/UTR and repetitive regions. We have also annotated piRNA clusters and have elucidated characteristic motifs within them. Further, expression profile for piRNAs in different tissues and from different developmental stages has been provided along with different disease systems which may help to understand the story behind the disease biology. Overall, piRNAQuest will serve as a useful resource for exploring piRNAome.
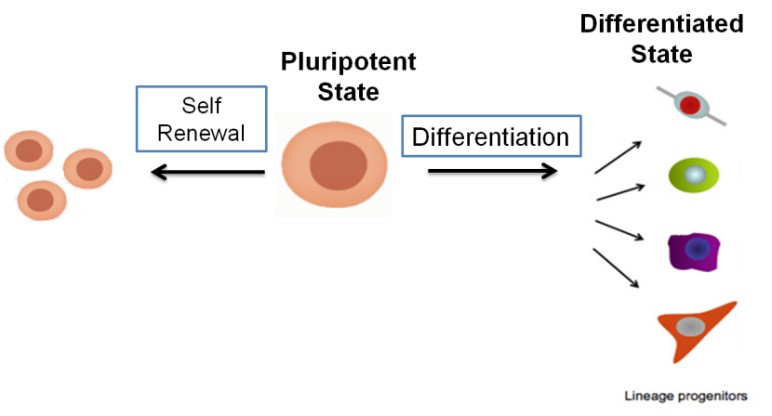
Induced pluripotent stem cells (iPSCs), generated from terminally differentiated cells, have the potential to self-renew and differentiate into cells corresponding to the three germ layers which make them an invaluable tool for regenerative medicine. However, the same properties make them oncogenic which can be transmitted to their derivatives too.
Epigenetic perturbations are likely to be the cause for transcriptional misregulation which induces oncogenicity within these derivatives. Regulatory noncoding RNAs (rncRNAs) play a role in epigenetic modifications. RncRNAs like miRNAs, have an important function towards controlling the chromatin modification through directly targeting epigenetic modifiers. We are trying to explore the role miRNAs towards inducing epigenetic changes within the stem cell derivatives corresponding to the three germ layers i.e. ectoderm, endoderm and mesoderm which makes them oncogenic and to reveal the mRNA-miRNA crosstalk which is responsible for the ‘oncogenic signature’ underlying these cells.
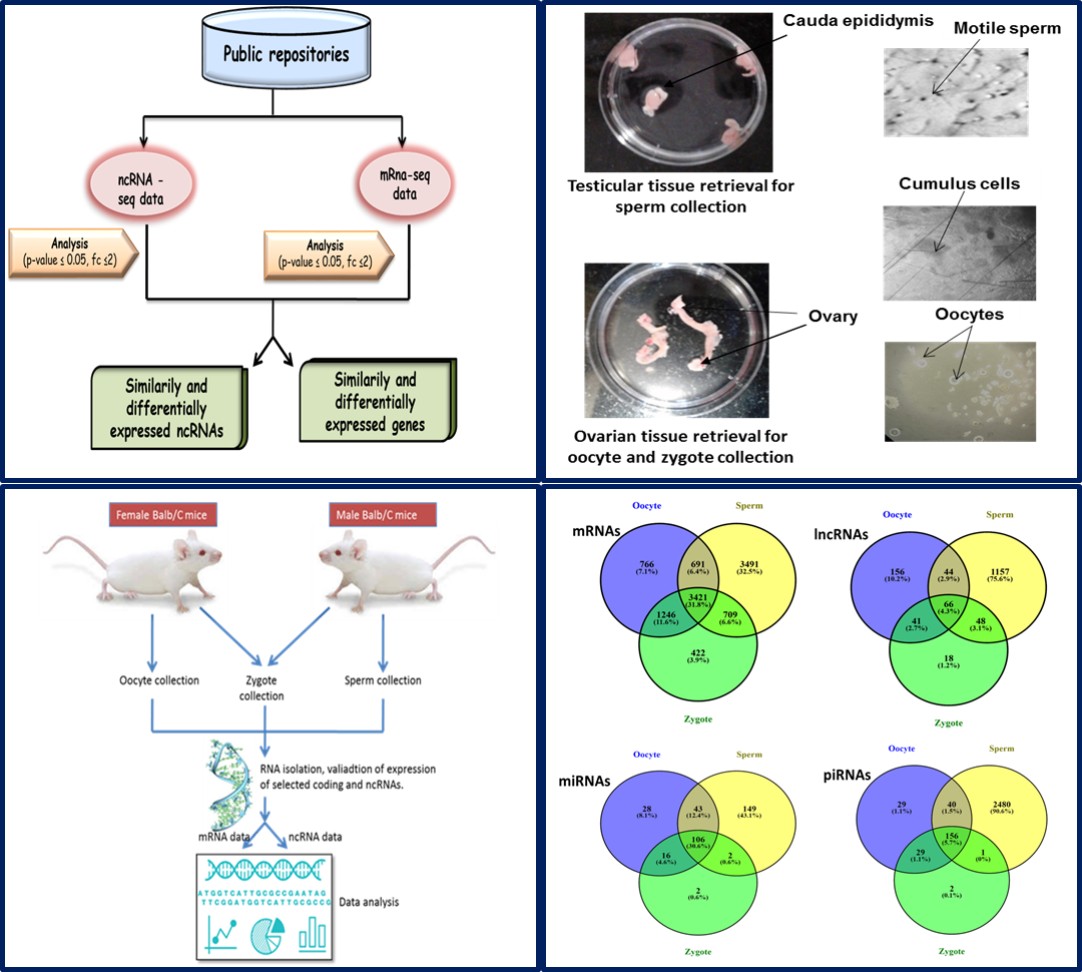
Regulatory RNAs viz; miRNAs, piRNAs and lncRNAs have been proposed to regulate many aspects of development including fertilization, zygotic genome activation and the maintenance of pluripotency during early prenatal development. Pluripotent stem cells derived from prenatal developmental stages have a tremendous potential for regenerative therapy and tissue engineering. However, it is absolutely necessary to study the complex gene regulation and reprogramming of these cells in different cell types before putting them for clinical use. In our lab we have screened 10 Transcription factors (TFs) (including novel markers viz; Notch3, Meis1, Gli3 and Srf) which are essential for maintenance, self-renewal and differentiation of pluripotency substates (Deb et al., 2017). Presently we are investigating the complex cross talk between protein coding genes and lncRNAs during neural lineage differentiation. Very recently we have also started studying the parental contribution of noncoding RNAs from sperm and oocytes to the newly formed zygote. This will help to understand fertilization and embryonic development events in more detail. Through this work we aim to find mRNA/ncRNA signatures’ which play key role in these developmental processes.
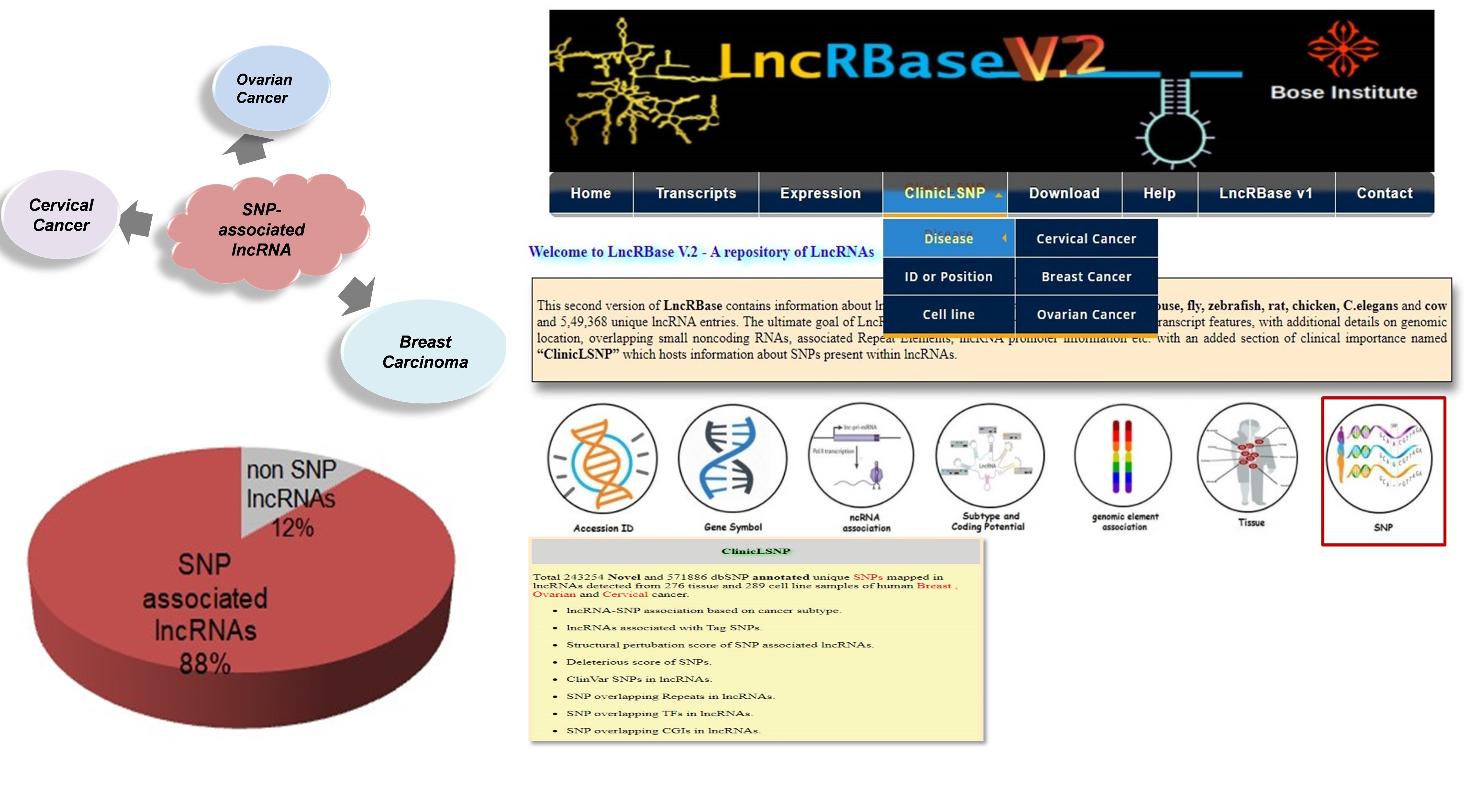
Long noncoding RNAs(lncRNAs) harboring SNPs or SNP-associated lncRNAs can have altered functions and can subsequently cause phenotypic changes within the system. We have focused on ovarian, breast and cervical cancer are reported to be most prevalent among Indian women. Shared genetic architecture leaves an open question as to whether they share a plexus of common cellular events guided by genetic variations within the genome. We aim to identify gene targets of these SNP-associated lncRNAs in female cancer systems. Identifying predisposing candidate SNP-associated lncRNAs and their target genes will help to identify high-risk groups which in turn will aid in early diagnosis and improve the survival rate of patients suffering from these highly prevalent malignancies.
In this connection we have developed a comprehensive resource for lncRNAs entitled “LncRBase” initially to study the various important features of lncRNAs which has been recently updated to provide a modified set of information regarding multispecies lncRNAs as well as genetic variants present within lncRNA loci.
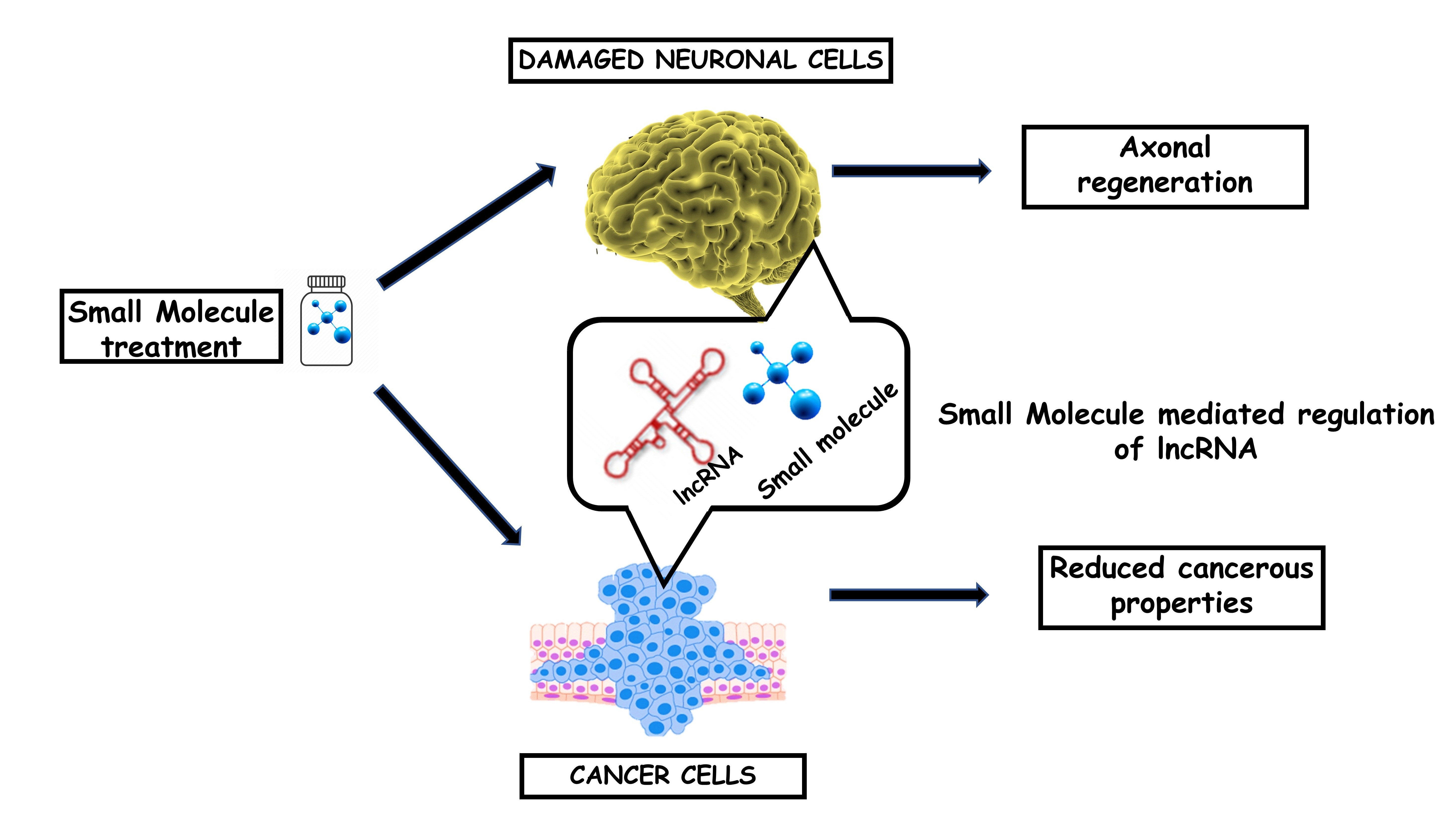
Recently, tremendous efforts and progress have been made in identifying novel factors and concepts in cancer biology as well as in neurobiology, where the protein-coding world represents only one side of the coin. LncRNA molecules have evolved as novel therapeutic agents in combating different diseases including cancer and different neurodegenerative diseases. While much emphasis has been centered on the role of lncRNAs in human disease, a gap remains in utilizing this knowledge to develop target specific approaches for disrupting lncRNA pathways for therapeutic gain. LncRNAs carry genomic signatures which can serve as a clue to establish the “connections” between drug and diseases. Our goal is to provide a better understanding of not only regarding the role of lncRNAs in the regulation of disease-relevant genes but also to utilize this knowledge in developing targeted therapeutics.