LncPTPred predicts the interaction probabilities between a given Long Non-Coding RNA (lncRNA) & a particular Protein. It extracts position specific binding location across the given lncRNA sequence.
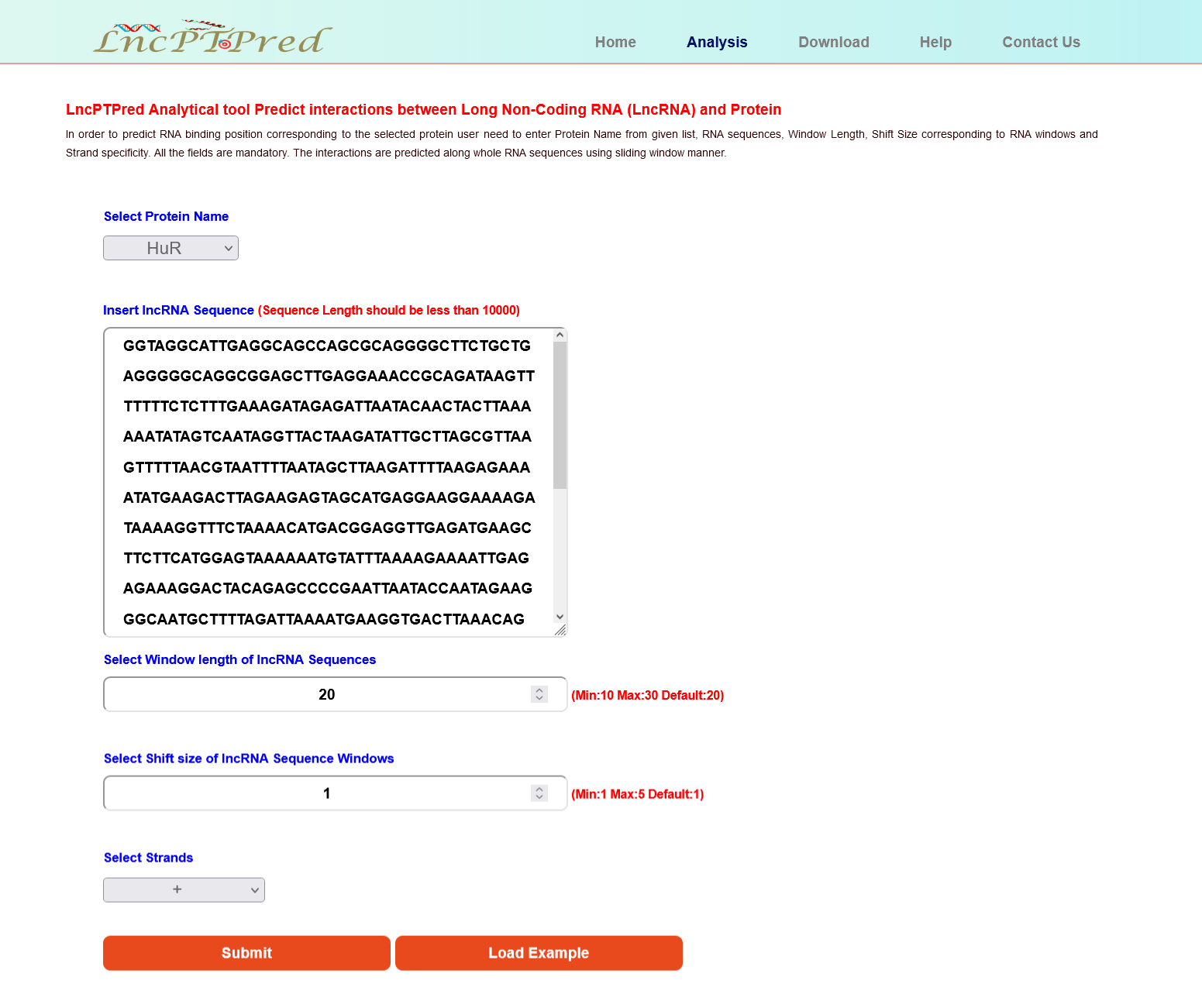
This tool extract position specific binding probabilities across the given lncRNA loci using a shifting window of specific size. In order to execute it, user need to enter the following:
- Select protein name from given list of input proteins.
- Enter the lncRNA sequence (Sequence Length should be less than 10000nt). For lncRNA sequence length > 10000 nts download the standalone version from Github.
- Select the shifting window length. Here max value is 40, min value is 10 and default is 20.
- Enter the shift size. Here max value is 5, min and default value is 1.
- Specify the strand + or - (Provide Exact Strand Information corresponding to the Input lncRNA Sequence otherwise it will generate wrong result).
- All the fields are mandatory.
- On clicking the Load Example an instance will be loaded. This can be run as demo by the user for the ease of using LncPTPred.
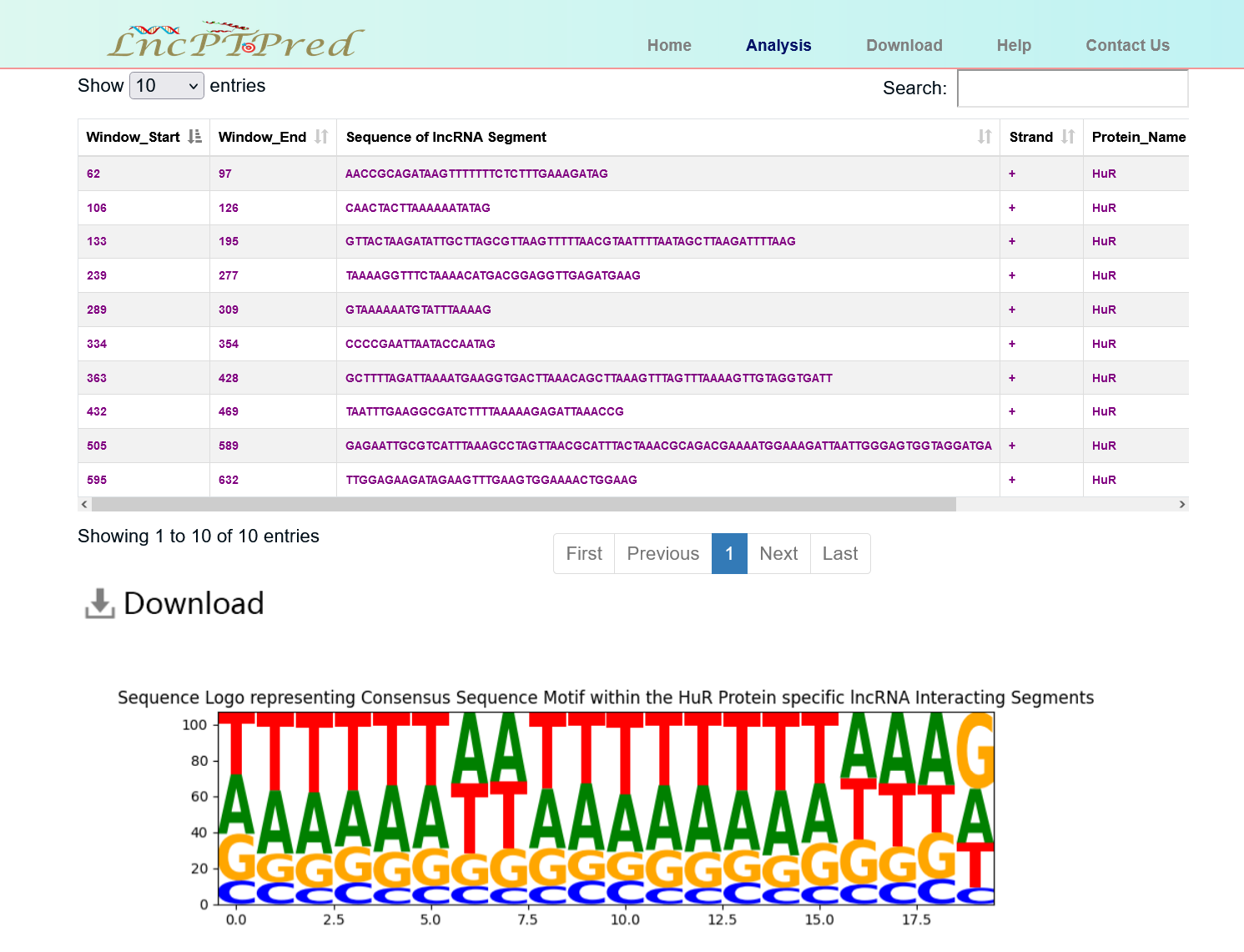
In the output page, the following will be obtained by the user: Prediction Result of interacting lncRNA sequence segments and the Motif plot.
- Data table containing specific location of binding lncRNA segments represented by start & end location, their corresponding sequences along with their interaction probabilities.
- Motif plot represents consensus sequence motif present within the interacting lncRNA segments corresponding to a particular protein.
- Users can download the whole prediction result by clicking the “Download” button.